 Molecular Basis of Calcitriol and Cell Signaling Pathways: Protective Effects Against Cancer Rachael Latshaw, Rachel Schaefer, Jeremy Bonor, Beth Bragdon and Anja Nohe University of Delaware Department of Biological Sciences Clinical research reveals an
inverse relationship between serum levels of calcitriol, the
biologically active form of vitamin D, and the risk of developing and
surviving hormone-dependent cancers. Calcitriol’s role as a nuclear
transcription factor allows it to regulate cell growth, proliferation,
differentiation and apoptosis. The mechanisms by which calcitriol
functions at the molecular level are unknown, leaving the reason this
molecule improves cancer prognoses a question mark among physicians and
scientists. With lacking knowledge on the relationship between cancer
and calcitriol signaling pathways and calcitriol’s absorbance into
cancerous and normal cells, research must focus on the mechanism of the
vitamin D receptor (VDR) and cellular uptake of calcitriol. To begin
this research, calcitriol must be successfully coupled to quantum dots
(QDs), high luminescence particles with a narrow emission spectrum. QDs
are fluorescent nanoparticles with a cadmium selenide core and zinc
sulfide shell used to tag calcitriol for live cell imaging. Infrared
analysis is used to verify coupling and show a shift where the
polyacrylic acid coating on QDs reacts with a hydroxide on calcitriol
to form an ester. A reporter gene assay confirms calcitriol is active
and confocal microscopy provides live cell imaging of calcitriol
absorbance through the plasma membrane of C2C12 mouse myoblastic stem
cells.
|
 Cloning and expression of neural cell adhesion molecules for functional studies of prostate cancer metastasis Nadia Lepori-Bui, Keith Jansson, and Robert A. Sikes Center for Translational Cancer Research, Laboratory for Cancer Ontogeny and Therapeutics Background:
Prostate cancer (PCa) is the most commonly diagnosed cancer in U.S.
males. Metastatic PCa results in over 27,000 deaths annually and
>85% involve metastasis to bone, particularly to the lumbar and
sacral spine. Current paradigms state that PCa metastasis is
through the circulation and/or lymphatic system, but this does not
explain the specific localization of PCa to bone and, more
specifically, the lumbar spine. We hypothesize that PCa
metastasizes to the spine via adhesion to nerve and/or nerve-associated
(Schwann) cells via the V-set immunoglobulin (Ig) domain of a novel
cell adhesion molecule, SCN2B. Schwann cells express a potential
cognate binding partner of SCN2B, called MPZL2 or Myelin-P0-like
protein. Tenascin-R (TnR) also is involved in neural tracking,
but has domains repulsive to SCN2B. Methods: Restriction enzymes were
used to clone MPZL2 and the repulsive domain of TnR into mammalian
expression vectors as fusion constructs with a small peptide tag
included for purification. Amplification of fragments for cloning was
accomplished using PCR-directed amplification, cloning into TA-vector
intermediates followed by subcloning those inserts into pcDNA 3.1
myc-His and PLN1 mammalian expression vectors. Results: The full length cDNA of
MPZL2-myc-His has been successfully subcloned and transfected into LN
cells. MPZL2-ectodomain and TnR were amplified successfully and
cloned into the TOPO vector but not yet into mammalian expression
vectors. Conclusions: The
cloning of these domains will allow adhesion studies with SCN2B and
other cellular adhesion molecules to determine their role in PCa
metastasis. Additionally, transfecting the full length isoforms
of these CAMs into the LNCAP progression model will allow us to
determine possible functional effects of constitutive
overexpression.
|
 Dietary fat may influence gut bacteria profile and diversity in Mus musculus Tianyu (Tom) Liu, Amy C. Vollmer, and Sarah Hiebert Burch Department of Biology, Swarthmore College Mammalian gut microbes have
been implicated in a variety of functions including the regulation of
energy intake and storage, the control of immune system development and
activity, and the synthesis of novel chemicals. Diet is a powerful
predictor of fecal bacteria profile across mammalian species, and
obesity has been correlated with shifts in gut bacterial profiles and a
decrease in bacterial diversity. The effect of oligosaccharides on gut
bacterial profiles has been well documented but the effect of dietary
fatty acid is poorly understood. Previously, groups of Mus musculus
were fed different diets rich in saturated fatty acids, omega-3
polyunsaturated fatty acids, and omega-6 polyunstaurated fatty acids
over a ten-week period. Mouse fecal pellets were collected before and
after the study. In our investigations, bacterial DNA was extracted
from the pellets and characterized by 16S rRNA analysis. Sections of
the hypervariable V3 region of bacterial 16S rRNA sequence were
amplified, cloned, sequenced, and classified to observe differences in
ratios of bacterial phytotypes as a result of differing dietary fatty
acid content. 16S rRNA sequences were also used to create phylogentic
trees and rarefaction curves to compare bacterial diversity among the
experimental groups. Results may elucidate the role of bacteria in how
diet affects the body and suggest dietary methods to regulate human
health.
|
|
Microbe-Mineral
Interactions and their Influence on Selenium Transformations in Soil
Environments
Allison M. Moran and Keka C. Biswas Wesley College Dover, DE 19901 Selenium occupies a unique
position in regards to its continuing conflicting aspects of its
toxicological and nutritional significance in bacterial reduction of
selenium (Se). Bacterial reduction of selenium (Se) oxyanions Se(VI)
and Se(IV) to elemental Se is one of the major biogeochemical processes
removing Se from agricultural drainage water and the depositing Se in
the sediment. Indigenous bacteria have a significant role in the
biogeochemical cycling of Se and may be stimulated by addition of a
suitable organic source for Se reduction. While few bacteria have the
capability of coupling the reduction of selenate or selenite to
elemental selenium, many aerobic bacteria have a glutathione-based
process of forming elemental selenium and several anaerobic bacteria
use metal reducing systems to produce elemental Selenium. Upon our
screening process, Se(VI)-and Se(IV)-reducing bacteria were
isolated from bedding litter and groundwater and identified by
amplification and sequencing of 16S rRNA. Bacillus strains appeared to
be dominant in the bacterial assemblages active in Se(VI) and Se(IV)
reduction. Using environmental bacterial isolates, we have followed the
reduction of selenite to colloidal elemental selenium by pure cultures
and the method detects elemental selenium even if elemental selenium is
formed inside the cells. Our research addresses a method for
measuring colloidal red selenium in the presence of chemicals typically
associated with soil or water containing toxic compounds. Using
environmental bacterial isolates, identified by colony PCR and DNA
sequencing, we have followed the reduction of selenite to colloidal
elemental selenium detected by AFM and Confocal microscopy techniques.
This Project described was supported by NSF-EPSCoR Grant EPS-0814251
obtained through Delaware Biotechnology Institute.
|
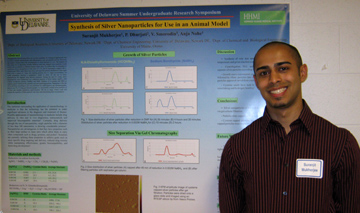 Synthesis of Silver Nanoparticles for Use in an Animal Model Suranjit Mukherjee, Prasad Dhurjati, Vladimir Smorodin, and Anja Nohe Department of Biological Sciences Nanoparticles derived from
silver are an ideal candidate for biological applications due to
silver’s natural anti-bacterial, anti-inflammatory, and anti-platelet
activity. However, not much is known about the accumulation of silver
particles in vivo. The purpose of this study was to determine whether
different sized and charged silver particles would selectively
accumulate in different tissues within mice. To achieve this we
attempted to synthesize silver nanoparticles in the sizes of 5, 10, and
20 nanometers capped with L-cysteine and MSA, inject them into mice,
and track the particle distribution at the organ level over a course of
5 days. By tracking particle size vs. organ accumulation, we could
potentially develop a quantitative model to allow for the use of silver
nanoparticles for targeted drug delivery purposes. During the past 10
weeks, silver nanoparticles were synthesized via a reduction reaction
of silver nitrate (AgNO3) with sodium borohyride solution (NaBH4) and
DMF. Addition of L-cysteine and MSA as capping agents stabilized the
particles and prevented aggregation. Separation of the the silver
particles according to size was performed by centrifugation, gel
chromatography, dialysis and TLC. Using gel chromatography, we
successfully isolated silver particles of 4-15nm and another population
approximately 100nm in range. Future work will include the injection of
these particles into mice and determining their accumulation. Funding
for this work was provided by HHMI.
|
 (First Place in the Sigma Xi Oral Presentations) Pathogenicity Islands Are Discrete and Ancient Integrative Elements: An Evolutionary and Functional Analysis Michael G. Napolitano, Salvador Almagro-Moreno, and E. Fidelma Boyd Department of Biological Sciences The evolution and emergence of
pathogenic bacteria is tightly linked to the horizontal transfer of
pathogenicity islands (PAIs) and bacteriophages encoding a range of
virulence factors. The current theoretical framework is that PAIs and
other genomic islands (GEIs) are a combination of diverse integrative
elements such as bacteriophages, integrative conjugative elements
(ICE), or plasmids. In this work, we describe how integrases from
phages and pathogenicity islands form distinct phylogenetic groups. In
addition, we show that island-encoded integrases are not evolutionarily
related to a range of integrative elements. These findings do not
support the view that pathogenicity and genomic islands are a broad
group of integrative elements derived from other elements. Using VPI-2
as a model for pathogenicity islands, we determined the role of the
VPI-2-encoded integrase (intVPI-2) and two putative excisionases (vefA
and vefB) in excision. We examined VPI-2 excision levels in V. cholerae
mutant strains (delta)intVPI-2, (delta)vefA and (delta)vefB and also in
strains that overexpressed these genes. We found negligible levels of
excision in the (delta)intVPI-2 strain whereas overexpression of
intVPI-2 showed a marked increase in excision. In the (delta)vefA or
(delta)vefB strains the excision levels of VPI-2 remained the same,
however, overexpression of vefB (VC1809) showed a 2-fold increase in
excision. Overexpression of vefA (VC1785) in the wildtype causes
complete excision of the island, however complimenting vefA with the
deleted strains (delta)vefA and (delta)vefB leads to only a minor
increase in excision above basal levels.
|
 Hyaluronic Acid Receptor Proteins and Their Roles in Prostate Cancer Metastasis Ngoc T. Nguyen, Lisa A. Gurski, and Mary C. Farach-Carson Department of Biological Sciences and the Center for Translational Cancer Research, University of Delaware Hyaluronic acid (HA) is a
polysaccharide that is distributed ubiquitously throughout the
extracellular matrix (ECM) of all tissues and is especially prevalent
in connective tissues such as bone and cartilage. HA plays a role in
cell motility and invasion, processes involved in metastasis, through
engagement with cell surface receptors, such as cluster of
differentiation 44 (CD44) and receptor for hyaluronan-mediated motility
(RHAMM). The purpose of this project is to study the expression of
these HA receptor proteins and to determine if more motile, invasive
cells show increased expression of HA receptors. We utilized the lymph
node-derived cancer of the prostate (LNCaP) progression model of human
prostate cancer (LNCaP, C-4, C4-2, and C4-2B), cell lines derived from
a common lineage that differ in their ability to form distant
metastases. This model progresses from non-metastatic LNCaP cells to
metastatic, highly aggressive C4-2B cells. Through Western blotting, we
have discovered that CD44 expression does not significantly increase
among the cells in the progression model, suggesting that invasiveness
and CD44 expression were not directly correlated in prostate cancer.
Western analysis did, however, demonstrate a significant increase in
RHAMM expression from LNCaP cells to the more invasive lines, conveying
that RHAMM expression was important for prostate cancer aggressiveness
and metastasis. Furthermore, we have employed immunofluorescence to
determine protein localization. This project is funded by the Howard
Hughes Medical Institute.
|
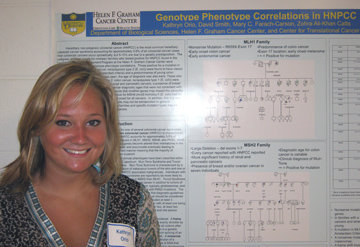 Genotype Phenotype Correlations in Hereditary Non-polyposis Colorectal Cancer Kathryn Orio, David Smith, Mary C. Farach-Carson, and Zohra Ali-Khan Catts Department of Biological Sciences, Helen F. Graham Cancer Center, and Center for Translational Cancer Research, Newark, DE. Hereditary non-polyposis
colorectal cancer (HNPCC) is the most common hereditary colorectal
cancer syndrome accounting for approximately 3-5% of all colorectal
cancer cases. Most colorectal cancers occur sporadically, but
5-10% are due to a genetic predisposition. The pedigrees and test
results for nineteen families who tested positive for HNPCC found in
the Familial Cancer Risk Assessment Program at the Helen F. Graham
Cancer Center were collected and analyzed for genotype phenotype
correlations. Those positive for a mutation in MLH1 (MutL homolog
1, colon cancer, nonpolyposis type 2 (E. coli)) were found to have
classic HNPCC (most families meeting Amsterdam criteria) and a
predominance of young colon cancers. When endometrial cancer was
seen, the age of diagnosis was also early. Those who tested positive
for MSH2 (mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli))
were found to have a more significant history of renal and pancreatic
cancers, a presence of breast and/or ovarian cancer, and variable colon
cancer diagnostic ages that were not consistent with mutation type or
location. One could hypothesize that modifier genes may impact
the variability seen within these families. Those testing
positive for MSH6 (mutS homolog 6 (E. coli)) had no predominance of
colon cancer and later age of onset for all cancers. In addition,
this is a very small sample of HNPCC families, so these results may not
be extrapolated to general population. Additional research
including more families and specific mutation types may be needed to
achieve statistically significant conclusions.
|
 P2 Receptors and ATP enhances the migration of prostate cancer cells Wachen Peters, Christine Maguire, Randall L. Duncan, and Robert A. Sikes Department of Biological Sciences Background:
Purinergic signaling stimulates many biological processes such as cell
proliferation, differentiation, and apoptosis. Two classes of
purinergic receptors, GPCR (P2Y) and gated channels (P2X), have been
identified that bind ATP as a ligand. Similarly, activation of
these receptors by ATP can promote cell migration, a critical component
of wound healing. ATP also has been shown to have an anticancer effect
on in vivo. Herein, we describe the effect of ATP on the
migration of an isogenic progression series of prostate cancer (PCa)
cell lines on three different extracellular matrices, Collagen I
(bone), Matrigel (basement membrane), and Schwann cell-derived (Nerve
support cells) in order to mimic growth on these common sites or routes
of metastasis. We hypothesize that ATP treatment activates
purinergic receptors that increases cell migration on all three
extracellular matrices. Methods:
RT-PCR was used to determine the mRNA expression profile of P2
receptors and ecto-nucleotidases. 0.1nM ATP or 2U/ml apyrase was added
to PCa cell lines cultured on three different matrices in vitro.
Migration was examined using wound healing assays by photomicroscopy
for four days, as well as RT-PCR to determine effect of interaction
with each ECM on the mRNA profile. Results: ATP increased the migration
of LNCaP cells on Collagen I to a higher degree than on either Schwann
and matrigel over vehicle alone. Peak wound closure occurred after 48
hours, while treatment of cells with apyrase alone inhibited cell
migration. Differential expression of P2 receptors and
ectonucleotidases was seen when comparing growth of cells on the
matrices. Conclusions:
ATP is necessary for PCa migration on collagen I, matrigel and Schwann
cell matrix. The differential expression of P2 receptors is
speculated to be responsible for variable wound healing on
extracellular matrices. Fund:
Department of Defense HBCU/MI Undergraduate Research Training Grant,
PC080950
|
 The SPAFF as a predictor of marital satisfaction Lauren C Pulinka, Rachael Greenberg, Brittany Moriarty, Elana Graber and Jean-Philippe Laurenceau Department of Psychology Previous research has shown
that the success of marriages can be predicted by examining positive
and negative emotions during conflict tasks (Gottman & Levenson
2000). The Specific Affect Coding System (SPAFF) was developed and
refined for coding a range of positive and negative affects predictive
of marital outcomes (Coan & Gottman 2007). Based on observing
marital interactions, Gottman and Levenson (2000) used SPAFF codes to
predict early divorce based on the presence of negative affect and
later divorce based on the absence of positive affect. The current
study utilized behavioral observation of couple interactions to examine
emotions that may be predictive of relationship outcomes (e.g.,
relationship satisfaction, divorce) two years later. Participants
consisted of 120 heterosexual newlywed couples (married < 6 months)
from Miami-Dade, Florida. Couples were videotaped participating in a
warm-up task, two conflict tasks, and a love task, which are in the
process of being coded using the SPAFF. They completed this process at
three time points. Most research of this kind has focused on
positive and negative affects in the context of conflict interactions,
whereas the current study will also focus on the presence of positive
and negative affects in a positive (i.e., love) task. We are
currently coding the love tasks of the first time point and expect to
replicate the previous findings.
|
 Osteoarthritis and Fruit Flies: Studying the role of heparan sulfate proteoglycans in essential and homologous signaling pathways Rachel Randell, Richard Wittmeyer, and Erica Selva Department of Biological Sciences In humans, degradation of articular cartilage leads to osteoarthritis. Here, the Drosophila wing imaginal disc is used to model the extracellular environment during signaling events required for the maintenance of articular cartilage. In both human articular cartilage and in the fly wing disc, the extracellular matrix contains heparan sulfate proteoglycans (HSPGs), which are known to modulate signaling pathways (including Wnt, Hedgehog and BMP). In Drosophila, HSPGs promote extracelluar ligand movement and are responsible for distinct patterns of signaling pathway activation in the wing disc. In humans, the same signaling pathways maintain cartilage homeostasis and integrity. To study the effects of HSPGs on signaling, flies have been generated in which wild type tissue and proteoglycan mutant “clones” lay side by side in the same wing disc. Immunostaining of these discs for signaling ligands and activation targets reveals the importance of HSPGs for these essential signaling pathways. Our results will determine the role of HSPGs with the hope of identifying targets for osteoarthritis therapeutics. |
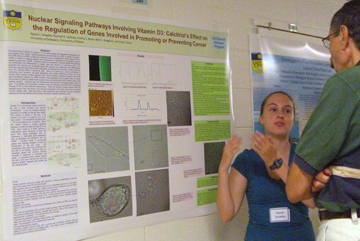 Nuclear Signaling Pathways Involving Vitamin D3: Calcitriol’s Effect on the Regulation of Genes Involved in Promoting or Preventing Cancer Rachel J. Schaefer, Anja G. Nohe, Rachael A. Latshaw, Jeremy C. Bonor, and Beth C. Bragdon1 University of Delaware, 1University of Maine The active form of vitamin D,
calcitriol or vitamin D3 regulates the expression of many genes in the
nucleus. Calcitriol enters the cell and binds to a VDR, or
vitamin D receptor. The VDR binds to a RXR, or Retinoid X
Receptor to form a heterodimer, which interacts with a VDRE, or vitamin
D response element with co-activators or repressors to up-regulate or
down-regulate a gene with a VDRE. Calcitriol controls the
expression of several genes that influence cell proliferation and
differentiation in healthy cells and prevent it in cancerous
cells. It is expected that calcitriol will up-regulate genes that
support cell proliferation and differentiation and down-regulate genes
that promote apoptosis, or programmed cell death in healthy cells and
have the opposite affect in cancerous cells. Since it is works as
a cancer preventative in controlling such functions, the gene
expression and nuclear pathways involved are being viewed using
calcitriol coupled quantum dots. Since calcitriol is present
throughout the entire gene expression pathway, the VDRE involved in the
expression can be viewed under a confocal microscope by labeling
calcitriol with quantum dots and allowing the C2C12 cells to take up
the labeled calcitriol. Quantum dots are nanoparticles that give
off a fluorescent glow under UV light. The stability of this
binding to calcitriol using the chemical cross linker DCC is measured
through FT-IR at three different temperatures. Once the
calcitriol coupled quantum dots are properly bound, they are used to
view the gene expression of the stem cells used to represent normal,
healthy cells. After the results of this experiment are
confirmed, the gene expression pathways involving calcitriol in the
nucleus will also be viewed in cancerous cells.
|
 The Effects of Histone Modifications on Lens Cell Denucleation Robert P. Sheehan, Yan Wang, and Melinda K. Duncan Department of Biological Sciences The lens of the eye contains
two different types of cells, epithelial cells and fiber cells, with
fibers cells deriving from the equatorial epithelium. When these cells
differentiate, cell organelles are broken down, including the nucleus.
However, small fragments of DNA from the nucleus remain. This led to
interest in studying this denucleation, to learn why these pieces of
DNA remained instead of being degraded. Our lab had previously studied
the dynamic role epigentic factors played in lens development, which
sparked the idea that similar epigenetic factors could be involved in
this denucleation. We thus began to study histone modifications in the
lens, specifically the trimethylation of Lysine 9 on histone H3. This
modification causes a compact, less accessible form of chromatin that
has no active gene transcription and may be less easily broken down. We
compared the presence of this modification to opposing ones such as the
acetylation of Lysine 9 on histone H3 and acetylation of Lysine 8 on
histone H4 which result in a loosely configured, accessible
euchromatin. In examining the lenses of humans and mice, we found a
pattern of expression of H3 Lys 9 trimetheylation in the transition
zone and fiber cells. This corresponded with fragments of DNA that were
expected as remnants of denucleation. However, there appeared to be no
acetylation of either histone studied. This suggests that remaining
fragments of the nuclei are entirely composed of heterochromatin, as
their highly compacted state prevented them from being broken down
along with the rest of the nucleus. Supported by Howard Hughes Medical
Institute and the National Eye Institute.
|
 Expression and localization of human sperm Plasma Membrane Calcium ATPase 4b (PMCA4b) which is required for normal motility in mouse sperm Katharine E Shelly, Rolands Aravindan, Patricia A Martin-DeLeon Department of Biological Sciences Sperm motility defects (asthenozoospermia) accounts for a large percentage of cases of male infertility. Two distinct types of motility are required of sperm to reach and fertilize an egg. Progressive motility assists sperm in reaching the fertilization site, while hyperactivated motility serves to free sperm from the lining of the oviductal wall in order to enable them to effect fertilization. While both are dependent on Ca2+ and ATP, the latter utilizes more ATP than progressive motility (1). In mice, hyperactivated motility and fertility are lost in the absence of Plasma Membrane Calcium ATPase isoform 4b (PMCA4b). This ATP-driven enzyme is the principal calcium efflux pump in sperm. Thus PMCA4b plays an important role in murine sperm function. The objective of this investigation is to characterize the presence of PMCA4b in human sperm by Western blotting, as well as to determine its subcellular localization by immunocytochemistry (ICC). Flow cytometric analysis will also be used to confirm the presence of PMCA4b. Detection of PMCA4b was attempted by Western blotting of solubilized proteins from 3 human samples using PMCA4 polyclonal antibody in concentrations of 1:500 and 1:833. Immunocytochemistry and flow cytometry were performed using various antibody concentrations (1:100, 1:250 and 1:500) for specific staining, compared to IgG controls for non-specific staining. This work was supported by the Charles Peter White Fellowship and the NIH-COBRE grant #5P20RR015588-07. |
 Possible Novel Mobile Genomic Elements and Vibrio Pathogenecity Islands in Vibrio mimicus Patrick Shock, Salvador Almagro-Moreno, Michael G. Napolitano, and E. Fidelma Boyd Department of Biological Sciences Vibrio cholerae is the causative agent of the diarrheal disease cholera. V. cholerae has known mobile genetic elements some of which are pathogenecity islands, such as VPI, VPI-2, VSP-I, and VSP-II, that may represent the initial genetic factor causing pandemic cholera. Moreover, another Vibrio species, V. mimicus has a very similar phenotype and genome as V. cholerae; however V. mimicus can only cause gastroenteritis, symptoms similar to non-pandemic V. cholerae. The similarity between the species phenotypes suggest the possibility that the pathogeneticy islands found in V. cholerae might transfer between these two genomes. Essentially, V. mimicus might act as a reservoir for the mobile genetic elements present in V. cholerae. Our first approach was to compare the genome of an environmental strain of V. mimicus MB451 to the genome of V. cholerae N16961. There were six regions in the V. mimicus MB451 genome that were not present in the V. cholerae N16961 genome. The six regions had G/C contents below the normal G/C content of 47.03 for V. mimicus MB451. Also, these regions encode an integrase, which suggest the possibility of horizontal gene transfer. The second approach was performing PCR assays on DNA of different V. mimicus strains to determine the presence of pathogenecity islands. In the PCR assay, two of three environmental strains and five of seven clinical strains of V. mimicus showed presence of VPI by positive PCR bands for the integrase (VC0847). |
 FTIRI and AFM analyses of cartilage and mineral tissues in HIP/RPL29-deficient mice Laura G. Sloofman, Padma P. Srinivasan, Elizabeth L. Adams, Kostas Verdelis, Lyudmila Spevak, Mary C. Farach-Carson, Adele L. Boskey, and Catherine B. Kirn-Safran Department of Biological Sciences Ribosomal proteins (RPs) are necessary for the maintenance of a normal protein synthetic rate. A short stature phenotype associated with increased bone fragility is observed in HIP/RPL29 null mice. We hypothesized that HIP/RPL29-null mutants exhibit changes in the relative amount of bone/cartilage constituents leading to structural imperfections at the molecular level. To understand the effects of HIP/RPL29 absence on mineralized tissue quality, microspectroscopy was performed on thin sections of growth plate and bone of tibiae in null and control mice of the C57BL6/J background. Using Fourier Transform Infrared Imaging (FTIRI), we showed that a decrease in collagen crosslinking during the development of HIP/RPL29 null bone precedes an overall enhancement in the relative extent of mineralization in adult bones and growth plate cartilage. Spectroscopic profiles and geometric data were also obtained for HIP/RPL29-null teeth, another mineralized tissue, via FTIRI and quantitative micro-CT. Like bone, HIP/RPL29-null teeth exhibited reduced geometric properties coupled with relative increases in the mineral densities of both dentin and enamel. Therefore HIP/RPL29 disruption induces defects in the relative distribution and/or structural organization of the mineral and organic phases, resulting in hypermineralization of bones and teeth. Cortical bone properties are being modeled using homogenization theory and elemental finite differences techniques. Additionally, we are developing a protocol to evaluate changes in the material property of articular cartilage in mutant mice using atomic force microscopy (AFM). Preliminary data suggest that HIP/RPL29 deletion does not alter articular cartilage elasticity. Because progenitor cells participating in articular cartilage formation are distinct from those contributing to underlying growth plate cartilage and shaft, it is possible that compensatory mechanisms exist in synovial joints and restore normal elasticity in articular cartilage of mutant bone. This project was funded by the Howard Hughes Medical Institute and COBRE NIH P20-RR016458. |
 The Role of the Unfolded Protein Response in Cataract Development of Connexin50 Mutants Jaime Stull, Melinda Duncan, and Zeynep Firtina Department of Biological Sciences Cataract is the leading cause
of blindness worldwide. Although the majority of cataracts are
age-related, they may also be caused by a heritable mutation. In humans
and mice, congenital cataract development occurs in Connexin50 (Cx50)
mutants. Our study involves two different Cx50 mutants, Cx50-S50P and
Cx50-G22R, that both express abnormal lens phenotypes- microphthalmia,
unorganized fiber cell structure, and cataract. While this observation
is not surprising since Cx50 is a gap junction component responsible
for supporting proper cell communication in the lens, it does not
explain why the observed lens abnormalities are more severe when
connexins are mutated compared to when the genes are absent in Cx50
null models. We hypothesized that when cells attempt to make Cx50 from
a mutated gene, the resulting protein does not fold properly, inducing
a cell-stress mechanism called the Unfolded Protein Response.
Upregulation of molecular folding chaperone BiP supported our proposal
of UPR activation in the lens and allowed for expansion of our
investigation to include heterozygous mutants. We found that at 13.5
dpc and 15.5 dpc, Cx50 homozygous mutants expressed high levels of UPR
activation, while Cx50 heterozygous mutants did not demonstrate a
substantial upregulation of UPR markers. Supported by Howard Hughes
Medical Institute, Fight for Sight, and the National Eye
Institute.
|
 Regulation of RNA under Abiotic Stress in Arabidopsis Matt Sullenberger, Linda Rymarquis, and Pamela Green Department of Plant and Soil Sciences, Delaware Biotechnology Institute, University of Delaware Plants
must deal with biotic and
abiotic stress on a daily basis in order to survive. One of the
ways
plants deal with stress is to alter the expression levels of microRNAs
(miRNAs). miRNAs are ~21 nucleotides RNA molecules, which regulate gene
expression by directing protein complexes to cleave messenger RNA
(mRNA)
“targets” in a sequence-specific manor (Sunkar et al. 2004).
This
research was designed to test the hypothesis that high temperature and
salinity
influence gene expression in plants by altering miRNA-mediated decay in
the
model plant Arabidopsis thaliana. To evaluate changes in
gene
expression, stresses were added to seedlings for 8 hours before being
harvested
for RNA extraction. A method known as Parallel Analysis of RNA
Ends
(PARE) was then used to capture 5’ monophosphorylated and
polyadenylated decay products
for deep sequencing. Analysis of the data potentially will reveal
novel
miRNA targets or known targets whose rate of miRNA-mediated decay is
altered
under stress conditions. Identifying novel or abiotic
stress-regulated
miRNA targets will add to the understanding of how plants respond to
abiotic
stress and lead to improvements in plant resistance. This work was
supported by
NSF EPSCoR, Grant EPS-0447610 and DOE (FGO02-91ER20021). |
 Protein Kinase C Mediates Purinergic Receptor Induced Contraction in MC3T3-E1 Osteoblasts Patricia Timothee, Victor Fomin, Kirk Czymmek and Randall L. Duncan Department of Biological Sciences Osteoblasts respond to
mechanical load with a rapid release of ATP that, in turn, binds to two
classes of purinergic receptors (P2 X and P2Y). Our lab has reported
that P2X7 receptor activation is essential to mechanotransduction in
osteoblasts. We have recently observed that activation of this receptor
results in a rapid change in osteoblast morphology and induces cellular
contraction. We hypothesize that activation of P2X7 receptors during
mechanical stimulation activates two distinct pathways, RhoA GTPase and
Protein Kinase C, that lead to the contraction of the osteoblast. Here,
we examined the changes in MC3T3-E1 preosteoblast morphology and
contraction using the Zeiss 5LIVE rapid confocal microscope during
activation of the P2X7 receptor and how these changes were affected by
inhibition of specific sites in the RhoA GTPase and PKC pathways.
BzATP, a known agonist of the P2X7 receptor, was added to
MC3T3-E1 cells and changes in cell area following BzATP stimulation
were quantitated using Differential Interphase Contrast (DIC)
microscopy. Addition of 0.5mM BzATP to MC3T3-E1 cells induced a
34.14% reduction in cell area while inhibition of the P2X7 receptor
with a blocking antibody significantly reduced the BzATP contraction.
These data suggest that the P2X7 receptor mediates this response.
Inhibition of PKC with the non-specific inhibitor, GF109203X,
attenuated the BzATP-induced contraction however blocking Rho Kinase
with LYXXX had little effect on BzATP induced contractions..
Finally, we were able to completely attenuate contraction by
removal of calcium from the extracellular media. This data
suggests an interactive role between PKC and the P2X7 receptor, as well
as the importance of extracellular calcium entry to potentiate
contraction. However, these data do not limit the potential role
of other P2X receptors during BzATP induced cell contraction.(supported
by INBRE 2P20RR016472-09 and NIH/NIDDK R01 DK058246).
|
 The Role of alg10 in N-glycosylation during Drosophila development Jessica Torres, Carly Dominica, Evan Lebois, and Erica Selva Department of Biolgical Sciences In order for proteins to
function properly in extracellular signaling, they may undergo several
post-translational modifications, such as Asparagine (N)
linked-glycosylation. Within this process, the alg10 gene encodes
a glycosyltransferase that adds the terminal glucose onto the
dolichol-oligosaccharide complex prior to its en masse transfer to
nascent polypeptides. In Drosophilia melanogaster, mutations in
alg10 are embryonic lethal, resulting in severe and pleiotropic
defects. Mutations in human homologs of other genes required to
build the dolichol-oligosaccharide complex result in a group of
disorders, congenital disorders of glycosylation, that cause infantile
death. This study aimed to determine the pathway through which
the alg10 mutation disrupts normal development. To achieve this
end, alg10 mutant embryos and imaginal eye discs were stained with
various molecular markers to define developmental defects in mutant
tissues. Indicators of the unfolded protein response (UPR) and of
apoptosis revealed elevated levels of both processes in alg10 mutant
tissues. Due to improper folding, proteins accumulate in the ER,
triggering UPR. The unfolded protein response, however, is not
able to accommodate the large excess of unfolded proteins and as a
result the cell undergoes apoptosis. This apoptotic mechanism
accounts for the rough eye phenotype observed in mutant eye discs and
perhaps the ventral open CNS phenotype observed in mutant
embryos. Continuing investigation into the CNS defects will be
focused on the specific pathway through which alg10 acts, with recent
data supporting a defect in axon pathfinding.
|
 |
Localization
of the Survival Motor Neuron and SIP1 in PC12 Cells
Amanda Vent1, Ilsa Gomez-Curet2, Wenlan Wang2,3 1University of Delaware, Newark DE; 2Nemours Biomedical Research, Alfred I. duPont Hospital for Children, Wilmington, DE; 3Department of Pediatrics, Thomas Jefferson University, Philadelphia, PA Spinal Muscular Atrophy (SMA)
is a neurological disease which occurs in 1 in 6000 live births caused
by deletions or mutations of the Survival Motor Neuron 1 (SMN) gene.
All patients have at least one copy of the highly homologous gene,
SMN2, which allows some function of motor neurons but ultimately does
not compensate for the loss of SMN1. The SMN protein often associates
with other proteins known as Gemins which aide in the assembly of small
nuclear ribonuclear proteins (snRNPs). This experiment studies the
localization of the SMN protein and one of these Gemins called the SMN
Interacting Protein 1 (SIP1), in the nucleus and cytoplasm of PC12
cells. Previous studies have shown a high degree of localization
between SMN and SIP1, as well as the loss of function of SIP1 upon
separation from SMN. Transfection of GFP was performed using Amaxa
Nucleofection, DharmaFECT Transfection, and Lipofectamine 2000 in order
to determine the most efficient method of transfection with the highest
cell viability. It was determined that the use of 1.5 μL DharmaFECT
reagent 4 was the most efficient method, however repeated experiment
failed to provide a successful transfection of differentiated PC12
cells. Lipofectamine 2000 and DharmaFECT reagents 1 and 3 were
ultimately used to transfect 293T and NIH 3T3 cells in order to observe
the localization of the fluorescent tagged wild-type and
patient-derived mutant plasmid constructs. / / Leptomycin B, an
antibiotic used to retain substrates within the nucleus which have been
marked for export by exportin1, was also used to study the localization
of SMN and SIP1 in the nucleus. Final results showed that 5-10 ng/mL of
Leptomycin B was unsuccessful in keeping the proteins in the nucleus of
naïve PC12 cells. A repeated experiment using differentiated PC12
cells and 20 ng/mL showed a localization of both SMN and SIP1 in the
nucleus.
|
 Biodegradation of Estrogenic Hormones by Poultry Litter Isolates Crystal J. Weesner and Keka C. Biswas Wesley College Dover, DE-1990 It is known that a variety of
bacteria have the unique capability to use sterols as a carbon source
to degrade both the steroid skeleton and the side-chain.
Characterization of these bacteria and their extra cellular enzymes
will have a wide range of applications, especially in studies involving
environmental degradation of veterinary antibiotics and steroidal
hormones. Research efforts were aimed at increasing our understanding
of the fate of endocrine destructing hormones from poultry excreta in
soil and subsurface environments. The goal was to develop predictive
models and guidelines related to land application of poultry manures to
agricultural lands. Elevated concentrations of estrogen [e.g. estradiol
and its primary metabolite, estrone] have been observed in soil,
groundwater, and surface waters impacted by runoff from agricultural
fields where manure has been applied [4]. The initial aim of this
undergraduate research project was the characterization and
identification of bacteria present in the bedding litter from
chicken farms that are capable of degrading manure-derived steroidal
estrogenic hormones. The second goal is to evaluate the
bacteria’s ability to then degrade steroid containing drugs. Bacterial
isolates from the chicken house and fresh litter were used to determine
degradation and hormone metabolism and degradation products were
analyzed by gas chromatography. / / This project described was
supported by NSF-EPSCoR RII, Grant Number EPS-0814251 obtained through
Delaware Biotechnology Institute. We would also like to acknowledge
NIH-NCRR-INBRE Grant number 2 P20 RR016472-09.
|
 Construction of the TOL-2 5’ 5Kb zCNR1p GAL4-VP16 plasmid and an investigation of its ability to assist in the visualization of the endocannibinoid system in a developing zebrafish embryo Maxwell Z. Wilson, Jonathan A. Raper, and Alison L. Dell University of Pennsylvania The development of an
organism’s nervous system is highly regulated by the widespread
utilization of G protein coupled receptors. Yet exploration into
the role of the endocannabinoid system, one of the most extensive
groups of G protein-coupled, neuromodulatory lipids and their
receptors, in development has hardly occurred. In this
investigation the cannabinoid receptor’s, abbreviated CB1, role in
zebrafish axon guidance was evaluated by genetically linking it to a
fluorescent protein, citrine, via the UAS-GAL4 system.
Preliminary results indicate that the 5 Kb upstream region to the CNR1
gene, which codes for the CB1 receptor, does in fact contain the
promoter for the CNR1 gene and that some expression of the CB1 receptor
is visible in the 24 and 48 hour developing zebrafish embryo.
|