Investigating the effects of TGF-β1 on the LNCaP Progression Model Within Bone Marrow Stroma Christopher W. Ahmer, Fayth L. Miles, Cindy Farach-Carson, and Robert A. Sikes Department of Biological Sciences Background.
Survival of prostate cancer (PCa) cells that have spread
to the bone marrow is the fatal step for cancer patients. Understanding
the interactions of metastatic prostate cancer cells with the normally
hostile bone marrow stromal cells in the bone environment is key to
controlling the disease. Transforming growth factor β1 (TGF-β1), a
cytokine that is highly elevated in the bone microenvironment, has a
prominent role in regulating cell growth, yet is strongly correlated
with advanced prostate cancer in vivo. Previously we have shown,
using the LNCaP progression model of genetically related, increasingly
metastatic human prostate cancer cells, that TGF-β1 promotes apoptosis
and inhibits cell growth. Additionally, we have shown that
conditioned medium (CM) from HS-5 bone marrow stromal cells originating
from progenitors surrounding the bone microvasculature strongly induces
apoptosis and neuroendocrine differentiation (NED) of prostate cancer
cells. Thus, we sought to determine if synergy existed between
HS-5 CM and TGF-B1 signaling, enhancing these effects. Methods. PCa cells were cultured
with HS-5 CM, as well as CM from HS-27a bone marrow stromal cells
derived from the endosteal surface of the bone, in the presence of
TGF-β1. Subsequently, live/dead assays were performed and
morphology examined. Results.
TGF-β1 in combination with HS-5 CM enhanced apoptosis of PCa cells as
compared to cells treated with HS-5 CM alone. Furthermore,
co-stimulation with TGF-β1 and HS-5 CM appeared to enhance NED,
characterized by increased process length and intercellular
extensions, as well as the NED marker, neuron specific enolase
(NSE). Conclusions.
these results highlight the significance of signaling in the activated
microenvironment in determining the fate of PCa cells during
colonization of bone. Summer Project was funded By the Stetson
Scholarship Program.
|
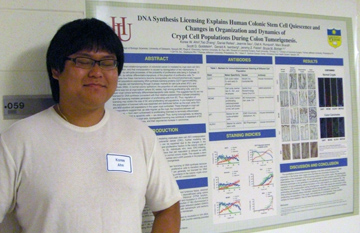 DNA synthesis licensing explains human colonic stem cell quiescence and changes in organization and dynamics of crypt cell populations during colon tumorigenesis Koree W. Ahn2 Tao Zhang1, Daniel Relles1, Jeannie Seu1, Olaf A. Runquist2, Marc Brandt3, Scott D. Goldstein4, Gerald A. Isenberg4, Jeremy Z. Fields5, Bruce M. Boman1,6,7 1 Dept of Biologic Sciences, University of Delaware, Newark DE; 2Dept of Chemistry, Hamline University, St. Paul, MN; 3Division of Colorectal Surgery, Rush University, Chicago, IL; 4 Division of Colorectal Surgery, Thomas Jefferson University, Philadelphia PA; 5 CA*TX, Inc., Gladwyne PA; 6 Thomas Jefferson University, Philadelphia, PA; 7 Helen F. Graham Cancer Center, Christiana Care Health System, Newark, DE We reported that
initiation/progression of colorectal cancer is mediated by crypt stem
cell (SC) overpopulation, and that overpopulation is caused by
dysregulation of two mechanisms: 1) regulation, by cell-cycle
processes, of the probability of proliferative cells being in S-phase;
2) regulation, by cellular differentiation/apoptosis, of the
proportion of proliferative cells. To investigate how these
mechanisms become dysregulated, we immunohistochemically mapped:
crypt cell populations expressing DNA synthesis licensing proteins
(CDT1/geminin/MCM2), which regulate cell transitioning through
S-phase; markers for cell-cycle arrest (P21); and apoptosis
(TUNEL). In normal colonic epithelium, the proportion of cells
expressing licensing proteins was low at crypt bottom (where SC
reside), high among proliferating cells, and nil in upper crypt (where
terminally differentiated/apoptotic cells reside). This suggests that
SC are not licensed for DNA synthesis (consistent with their
relative quiescence/low S-phase probability) and that licensing
mediates generation of proliferating cells from SC. Thus, regulation
of / licensing may control the size of SC and proliferating cell
populations. In pre-malignant crypts, the population of licensed
cells was expanded and distributed farther up the crypt, while
P21- and TUNEL-positive cell populations in the upper crypt
contracted. These changes in crypt cell subpopulations suggest
that, as cells migrate up the crypt, the transitions between cell
phenotypes — from stem (unlicensed) to proliferating (licensed) to
terminally-differentiated (cell-cycle arrested) to apoptotic
cells — are delayed. Thus, during tumorigenesis, by delaying cell
maturation along the crypt axis, dysregulated licensing may contribute
to expansion of SC and proliferating cell populations, and their
exponential increase in carcinomas.
|
|
Methylation
analysis of the G-C Rich DMPK gene:
Causation for the severity difference between adult-onset and congenital myotonic dystrophy? Brian Barnette, Sarah Swain, Susan Kirwin, and Vicky Funanage Alfred I. duPont Hospital for Children/Nemours Children’s Clinic Myotonic dystrophy type I is an
autosomal dominant genetic disease. The disease arises from an
expansion of a trinucleotide repeat, (CTG)n located in the 3’
untranslated (3’ UTR) region of the dystrophica myotonin protein kinase
(DMPK) gene on chromosome 19. Located proximal to this region is
a CpG island; a region of DNA with a high concentration of CpG
sequences that often associate with gene promoters in humans. The
purpose of this study was to determine the methylation status of this
CpG island in individuals affected with DM1, notably congenital DM1,
and to compare this methylation pattern with that seen in
individuals not affected by DM1. The method of
bisulfite-conversion was used to ascertain the methylation status of
the CpG island. This method converts all unmethylated cytosine
residues into uracil residues. The converted DNA is then
amplified by means of PCR where the converted uracil residues are
amplified as thymine residues. This amplified DNA is then cloned
and sequenced to compare the methylation status of the CpG island among
individuals. Currently, we are amplifying and sequencing this CpG
island. These techniques will be used to explore a relation, if any,
between methylation of the CpG island and the difference in expression
of full-length DMPK transcript between adult-onset and congenital DM.
|
 The role of Bcl-2 during neuronal migration and survival in the chick midbrain Cory D. Bovenzi and Deni S. Galileo Department of Biological Sciences, University of Delaware In the developing brain,
neurons are born in the ventricular zone and migrate outward via radial
glia scaffolds. There also is an early period of widespread programmed
cell death (PCD) in the brain. We believe there is a relation between
these two processes whereby the cells that die do so because of
anoikis: loss of contact with the necessary extracellular matrix
produced by radial glia. Remaining neurons are those that attached to
radial glial matrix and received survival signals through an
intracellular signaling pathway. In the developing chicken midbrain, we
believe this pathway involves fibronectin secreted by radial glia
interacting with the α8β1 integrin receptors on the neuronal membrane,
initiating a Focal Adhesion Kinase-mediated cascade eventually
increasing expression of Bcl-2, an anti-apoptotic protein. Higher
levels of Bcl-2 increase the likelihood that a cell will survive
periods of widespread PCD. To study the role of Bcl-2 in brain
development, this protein was ectopically expressed with a
replication-incompetent retroviral vector in the embryonic chick optic
tectum. An infected progenitor cell divides to produce a cohort of
clonal progeny that all carry the marker lacZ gene. The number and
distribution of lacZ + neurons in Bcl-2 overexpressing cohorts were
counted and compared to control clones. Expression of another potential
integrin involved (α5β1), whether contact with radial glia in culture
facilitates neuronal survival, and whether greater Bcl-2 normally has
an inverse relationship with cell death in the avian optic tectum were
also investigated. This project was funded by the Howard Hughes
Medical Institute
|
 MicroRNAs Associated with Environmental Stress in Arabidopsis (Second place oral presentation in Sigma Xi competition) Rebecca S. H. Brown, Dong-Hoon Jeong, and Pamela J. Green Department of Plant and Soil Science Plants are limited in the number of
ways that they can respond to environmental stress because they are
sessile organisms. Because of this, plants have developed
sophisticated mechanisms that enable them to cope with adverse
environmental conditions. One way plants respond to environmental
stress is by modifying their gene expression through the use of
microRNAs (miRNAs). miRNAs are noncoding small RNAs that regulate
gene expression at the posttranscriptional level by base pairing with
complementary messenger RNA (mRNA) molecules, causing either mRNA
cleavage or translational inhibition. To elucidate the roles of
miRNAs in environmental stress responses, wild type plants and miRNA
enriched mutant plants were subjected to various environmental
stresses, such as drought, cold, submergence, salt, and combined
stresses of submergence and salt. Small RNA libraries from
stress-treated seedlings and flowers were constructed and sequenced
using deep sequencing technologies. Computational analysis
revealed differential expression of miRNAs between stress-treated
plants and control plants with some miRNAs upregulated and some
downreguled by environmental stress. Additional analysis revealed
potentially novel miRNAs in Arabidopsis as well. These results
suggest that miRNAs play important roles in stress responses, and that
miRNA identification in Arabidopsis has not been saturated.
Future work will involve validating highly stress-regulated miRNAs and
prospective new miRNAs, and examining their potential target genes for
regulation. Stress-regulated miRNAs will be incorporated into
multi-network models and their biological roles will be hypothesized
and tested through functional studies. R.S.H.B. was supported by
the Howard Hughes Medical Institute, and NSF grant MCB#0548569 to
P.J.G. provided research support.
|
|
Molecular
Characterization, Isolation, and Identification of Chicken Litter
Bacteria
Shannon E. Carter1 , Thomas E. Hanson 2, and Keka C. Biswas1 1Wesley College, Dover DE 19901, 2 University of Delaware, Newark DE 19716 The initial aim of this
undergraduate research project was characterization and identification
of bacteria present in the bedding litter from chicken farms that are
capable of degrading manure-derived antibiotics and steroidal
estrogenic hormones like 17-β estradiol and estrone.
characterization of bacterial isolates through DNA extraction, colony
PCR , sequencing to understand molecular mechanisms using enzyme
isolation and protein purification techniques. Characterization of
these bacteria and their extra cellular enzymes will have a wide range
of applications, especially in studies involving environmental
degradation of veterinary antibiotics and steroidal hormones. In this
study, 16S rRNA sequencing was used to characterize bacterial isolates
from chicken litter. The use of 16S rRNA sequencing to accurately
identify bacterial isolates has become an essential operation in both
clinical and environmental microbiology laboratories. There are many
advantages in using the 16S rRNA sequencing including its presence in
almost all bacterial genomes, and because the function of this gene
over time has not changed. The second goal was to evaluate the
bacteria’s ability to then degrade estrogenic hormone and steroid
containing pharmaceutical drugs. This is to understand the
microbiological degradation process for the fission of the steroid
skeleton in the drugs and to document any relationship between the
degradation process and the presence of susceptible points (viz.
functional groups, double bonds, side chains, etc.). This project
is supported by NSF-EPSCoR Grant EPS-0814251, NIH NCRR INBRE grant 2
P20 RR016472-08 and The Delaware Biotechnology Institute.
|
 Function and Expression of Compatible Solute Transporters in Vibrio parahaemolyticus Nikhil B. Chinmaya, Seth Blumerman, & E. Fidelma Boyd Department of Biological Sciences Bacteria living in varying salt concentrations are under constant danger of death from the loss of the delicate osmotic equilibrium within their cells. Vibrio parahaemolyticus is a moderate halophile that is an emerging human pathogen known to cause gastroenteritis. V. parahaemolyticus lives in estuaries and oceans around the world in wide ranging salinities and osmotic conditions. Like most bacteria, V. parahaemolyticus has evolved a system of osmoadaptation that uptakes compatible solutes to protect and preserve the enzymatic activity of the bacterial cell. V. parahaemolyticus has four single component compatible solute transporters we took the previously cloned genes of these transporters designated VP1456, VP1723, VP1905 all located on chromosome I, and VPA0356 located on chromosome II. Whose function we have little to no understanding of in the bacterial cell. Therefore, to understand the function of these transporters We examined their function by cloning the genes and transforming them into pBBR1MCS creating pVP1456, pVP1723, pVP1905, and pVPA0356, a vector in a E. coli strain MKH13 (Transporter deficient strain of E. coli). We determined that some of these transporters are able to uptake glycine betaine but were not able to take up proline. To get a comprehensive understanding of the compatible solute transporters, and how they behave in their natural environment we also tested V. parahaemolyticus under specific conditions to observe the levels of gene expression of the particular transporter genes, to see a correlation. |
 Probing the Unknown: Development and Testing of PCR Assays for Viral Genes in Environmental Samples Jennifer E. Clarke1, Mara Hyatt2, William Kress, Rachel Marine, Michael Dumas, Sanchita Jamindar, Shawn Polson, K. Eric Wommack 1Linconln University, Lincoln University PA, 2Delaware Technical and Community College Polymerase chain reaction (PCR)
enables researchers to produce millions of copies of a specific DNA
sequence from minute quantities of a starting template. Applied to
environmental samples, PCR provides a means to access and explore the
diversity of genes within viruses and microorganisms. The central goal
of this project was to explore the environmental occurrence and
diversity of predicted open reading frames (ORFs) from two single
stranded DNA (ssDNA) viral metagenome libraries. Prior to development
of PCR assays, predicted ORFs were clustered by homology, and ranked
according to the number of member ORFs. Primer sets were
developed for three of the top twenty ORF clusters. A four step
procedure was used in the development of the assays. First, primer sets
were tested against a sample known to contain the gene of interest.
Second, temperature gradient PCR was used to discern the optimal
annealing temperature (Tm °C) for each of the primer sets. In each
case the optimal Tm°C was higher than the theoretical prediction.
Third, a positive PCR control was developed by isolating DNA of a
specific PCR amplicon from a Chesapeake Bay sample. Finally, a serial
dilution of the positive control DNA was tested against each
corresponding primer set. Testing of the primer sets against samples
from White Clay Creek and soil from the Newark Agricultural Experiment
Station revealed negative results even after the samples were spiked
with a positive control, suggesting inhibition of the PCR by the
environmental sample. For the soil samples it is hypothesized that
humic acid is causing this inhibition. This research was funded by the
National Science Foundation.
|
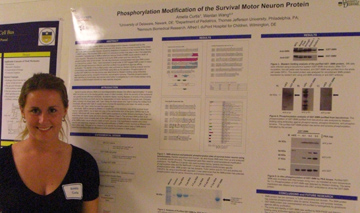 Phosphorylation Modification of the Survival Motor Neuron Protein Amelia Curtis and Wenlan Wang Nemours Biomedical Research, Alfred I. duPont Hospital for Children, Wilmington, DE Department of Pediatrics, Thomas Jefferson University, Philadelphia, PA Spinal muscular atrophy (SMA)
is a neurodegenerative disease characterized by the degeneration of
motor neurons in the spinal cord and atrophy of the proximal muscles of
the limbs and trunk. SMA is a genetic disease caused by deletions or
mutations in the Survival Motor Neuron 1 (SMN1) gene. The encoded SMN
protein is a ubiquitous protein that plays a role in the assembly of
snRNPs. Complete loss of SMN is lethal but reduced levels of SMN cause
selective death of motor neurons. The goal of my research is to purify
SMN protein and then identify the phosphorylation sites of this
protein. Our lab has previously overexpressed wild type SMN protein in
a baculovirus system. Here, I have expressed a small scale of
GST-tagged SMN protein in insect Sf9 cells through baculovirus
infection and then purified it by glutathione-agarose affinity
purification. The identity of the purified protein was confirmed by
Western blotting analysis and the concentration was determined by
SDS-PAGE using BSA as the standard. Phosphorylation of purified GST-SMN
from the baculovirus was determined by Western blotting using
antibodies against phosphor-serine, phosphor-threonine, and
phosphor-tyrosine. Potential phosphorylation sites identified by
bioinfomatic approaches were further investigated by in vitro kinase
assays using lysates or purified kinases followed by mass spectral
analyses.
|
 Characterization of Synovial Mesenchymal Stem Cells by Immunohistochemistry, Cell Differentiation, and Cytokine Level Analysis Belynda Dalecki, Revital Herrmann, and Paul Fawcett1 1Nemours Biomedical Research, A.I. duPont Hospital for Children Juvenile Idiopathic Arthritis
(JIA) is an autoimmune disease characterized by inflammation of joints
in pediatric patients under the age of 18. Lyme disease is caused by a
bacteria, Borrelia burgdorferi, carried by deer ticks and can be
classified as Lyme Acute (LA), with duration of symptoms less than 6
months, or Lyme Chronic (LC), with duration of symptoms longer than 6
months. Lyme Chronic arthritis occurs in about 10% of pediatric
patients who, despite treatment with antibiotics, continue to show
symptoms of inflammation. Synovial fluids were obtained for
analysis from patients undergoing arthrocentesis as part of their
clinical care. Cells were centrifuged, counted and morphologically
identified as leukocytes (neutrophils, lymphocytes, and monocytes).
Other small round cells in the fluid that, unlike leukocytes, have
exhibited an ability to grow and proliferate in culture were previously
identified as synoviocytes. In this study, we wanted to determine
whether the synoviocytes are mesenchymal stem cells (MSCs). MSCs have
the capability to differentiate into several cell lineages including
bone, fat, cartilage, and muscle. Using a sample obtained from a LC
patient, we tested the synoviocytes’ ability to undergo transformation
through Osteogenesis and Adipogenesis. Transformation was followed by
Enzyme-linked immunosorbent assay (ELISA) (R&D Systems) and protein
micro array (RayBiotech) to determine the transformation effect on
cytokine production by the cells. Finally, Immunohistochemistry was
used to identify mesenchymal stem cell markers on cultured cells
obtained from JIA, LA, and LC patients. This project was supported by
grant number 2 P20 RR016472-09 under the INBRE Program of the National
Center for Research Resources (NCRR), National Institutes of Health
(NIH).
|
 The Unwinding Efficiency of Simian Virus 40 T Antigen is affected by the Amount of Core Origin DNA Present Kyle Damken, Weiping Wang, and Daniel Simmons Department of Biological Sciences Simian Virus 40 (SV40) is a
small tumor virus that serves as an excellent experimental model for
eukaryotic DNA replication. T Antigen (T Ag) is a virally encoded
protein necessary for the initiation of SV40 DNA replication at its
single core origin. Certain host cellular proteins bind to T Ag,
forming the initiation complex and allowing DNA replication to
begin. The SV40 core origin (64 bp) consists of 3 distinct
regions, the central region is called Site II, where T Ag binds, the
early palindrome (EP) region, where the DNA is melted and the AT rich
track, where the DNA is untwisted. The purpose of this research
project was to determine which sequences of the core origin are
important for DNA unwinding. A series of unwinding assays were
performed using varying lengths of P-32 labeled origin DNA containing
different amounts of the EP region. The results showed that T Ag
was efficient at unwinding DNA with just Site II, but as base pairs
from EP were added unwinding activity dropped significantly.
Unwinding was restored when the entire EP region was present.
These data can be explained by a model describing the mechanism of
unwinding by T Ag. To compare these results with the unwinding of
circular DNA, DNA minicircles will be generated containing different
amounts of the core origin, and will be subjected to unwinding assays
with T Ag and Topo 1 to determine the importance of the EP region and
the AT tract for efficient SV40 DNA unwinding. This research will
help to show more clearly which parts of the core origin are needed for
DNA unwinding.
|
|
Investigating
the function and development of Motor Neurons In Wild Type and SMA
Spinal Cord Models
Bianca DeBroux, Rashida Williams, and Melissa Harrington Delaware State University, Biological Sciences Electrophysiology, Calcium
imaging, and Genotyping were used to measure and understand the
development of motor neurons in both normal and SMA spinal cord models.
These experiments used two mouse models one of which had the SMA mutant
gene and the other which did not. Electrophysiology with the Med 64
multielectrode recording system measure the neural activity within a
network of motor neurons. Calcium imaging was used to measure
intracellular communication, which supports the results with the Med 64
dishes that measure electrical motor neuron activity. The last
technique used was genotyping of the different mouse models. Knowing
the genotype of these model helps differentiate between the 3 different
type of mouse pups that we can obtain in a litter. Knowing specially if
these mice are homozygous wild type or homozygous for SMN knockout or
heterozygous will allow us to relate the exact difference between the
mutant and wild type model motor neuron activity to genotype. This
experiments thus far have allow us to obtain electrical signal on med
64 dishes, we were not yet able to determine the significance of motor
neuron activity recorded. With genotyping we saw bands for both the
mutant and wild type gene products.
|
 A Comparative Analysis of the Activation of RhoA and RhoC GTPase and Subsequent Cellular Activity DiSabatino, S. and Kenneth van Golen Department of Biological Sciences RhoA and RhoC are GTPase
enzymes that function as molecular switches within the cell that
control the actin cytoskeleton. When bound to GTP, RhoA and -C are in
an active state and are able to participate in various signal
transduction pathways within a stimulated cell. One of the main actions
of active RhoA and -C is the triggering of cytoskeletal rearrangement,
a process readily observed within invasive and motile cancer cells.
Considering RhoA and -C are over 90% homologous in structure at the
protein level, it has been assumed that they function identically. Data
from our laboratory and others demonstrates this is not case. The two
GTPases could potentially display temporal and/or spatial differences
in their activations. Using mouse fibroblast NIH3T3 cells as a model,
we hope to distinguish which membrane receptors, G-protein coupled
receptors or tyrosine kinase receptors, are predominantly responsible
for the activation of RhoA and RhoC and in what timeframe these
respective activations occur. The data gathered from these experiments
will provide a more specific knowledge of the relationship between RhoA
and RhoC as they relate to cell motility and potentially to cancer
progression. The practical applications of such knowledge could be
potentially far reaching in both the research and medical communities.
This summer’s work has been funded by the Charles Peter White Grant.
|
 Efflux Pump Upregulation in Salmonella Resistant to Dodecyltrimethylammonium Chloride Aleksey Dvorzhinskiy, Megan Kautz, Natalie Stevenson, and Diane Herson Department of Biological Sciences Salmonella spp. are
gram-negative pathogens estimated to cause 1.4 million cases of food
poisoning annually in the United States and are increasingly becoming
resistant to disinfection in natural, clinical, and laboratory
environments. In this study Salmonella enterica 4931 strains were
developed which were able to grow in the presence of 500-600 ppm
Dodecyltrimethylammonium chloride (DTAC), a quaternary ammonium
compound. These strains with reduced susceptibility to DTAC (SRS)
also showed a cross resistance to penicillin. Later, the SRS
strains were passaged in the absence of DTAC to mimic environmental
conditions after exposure to DTAC. The minimum inhibitory concentration
(MIC) of DTAC for these DSRS strains did not vary appreciably from the
parental level for fifty passages. Reduced susceptibility to DTAC has
been reported to be correlated with an increase in the amount of
transcription of acrB, an efflux pump which has been linked to
multi-drug resistance. The results from our research support this
finding. This study showed that all SRS and DSRS strains in the
presence of the efflux pump inhibitor Carbonyl cyanide
3-chlorophenylhydrazone (CCCP) showed a greater sensitivity to DTAC
than the parental strain. This is possibly another physiological
indication of a greater reliance of all resistant strains on efflux
pumps for defense against anti-microbial compounds. The discovery of
efflux pump inhibitors such as CCCP could lead to advances in combating
SRS strains in clinical and natural environments. This research was
generously funded by the Howard Hughes Medical Institute.
|
 Characterization of Adult Human Neural Progenitor Cell Differentiation In Vitro and In Vivo Michele Fascelli, Deni S. Galileo, and Grace Yang Department of Biolgical Sciences Adult human neural progenitor
cells (AHNPs) can be isolated from the adult hippocampus and cerebral
cortex. Others showed they can differentiate into neurons and
astrocytes in vitro and when injected into mouse brains, while our lab
had similar results in chick embryo brains. AHNP studies have
future applications for regenerative medicine and understanding brain
tumor carcinogenesis. I injected GFP-expressing AHNPs into E5
chick embryo midbrain and hindbrain ventricles in order to visualize
their motility and development. Approximately 10 days after
injection, brains were dissected, sectioned at 200 microns on a
vibratome, and screened for green cells using a dissecting microscope
with epifluorescence. Sections will be immunostained for cell
type-specific markers and analyzed by confocal microscopy.
Currently, different methods are being evaluated for facilitating
antibody penetration into the sections. Western blot analysis
showed full-length L1 protein expression in AHNP cells;
immunocytochemistry confirmed the expression of membrane surface L1 (L1
is a transmembrane protein that has a different function when
proteolyzed from the surface). Pax-6 transcription factor also
was detected by western blot analysis; however, immunostaining for
Pax-6 did not reveal expected nuclear localization. Future work
will be done with co-cultures of AHNPs and chick brain cells as an in
vitro¬ model for AHNP cell differentiation. These will be
characterized for differentiated neurons, astrocytes, and
oligodendrocytes. This project was funded by the Howard Hughes
Medical Institute.
|
 Abnormal Protein Expression in SMA Motor Neurons derived from Embryonic Stem Cells Steven Foltz1, Dosh Whye2, and Wenlan Wang2,3 1University of Delaware, Newark, DE, 2Nemours Biomedical Research, AI DuPont Hospital for Children, Wilmington, DE; 3Department of Pediatrics, Thomas Jefferson University, Philadelphia, PA Spinal Muscular Atrophy (SMA)
is a neuromuscular disease characterized by degeneration of spinal
motor neurons accompanied by muscle paralysis. Deletions or
mutations of the survival motor neuron 1 (SMN1) gene have been cited as
the cause of this devastating genetic disorder. Despite the
highly specific tissue degeneration observed in SMA patients, the SMN
protein encoded by the SMN1 gene is ubiquitously expressed. Thus
far, conclusive evidence as to why loss of a ubiquitously expressed
protein would result in the highly selective loss of motor neurons
remains to be established. Here, murine embryonic stem cells
derived from control and SMA mice were differentiated into motor
neurons by retinoic acid and small molecule agonist of sonic
hedgehog. Abnormal protein expression in SMA motor neurons was
analyzed by proteomic and Western blotting analyses. These studies
revealed that several stress-related proteins are upregulated in SMA
motor neurons differentiated from embryonic stem cells. Current
work is devoted to elucidating the specific pathway(s) that are
affected in SMA motor neurons, with the ultimate goal of identifying
SMA biomarkers that can be used in clinical trials and the revelation
of pathways that can be regulated to treat SMA.
|
 The effect of 1H7 and 3B7 treatment on molecular signaling through the IGF-IR in prostate cancer Brett Friedberg, Erica Dashner, and Kenneth L. van Golen Department of Biological Sciences Prostate Cancer (PCa) affects
one out of every six American men and is the second leading cause of
cancer related deaths among males. Advanced PCa easily
metastasizes to bone which causes unrelenting pain. Current
chemotherapies do not help in advanced PCa. The humanized antibody,
1H7, has been used in other cancers to reduce tumor size but it has not
been tested in PCa. 1H7 and another antibody, 3B7, binds the growth-
and metastasis-promoting Insulin-like Growth Factor 1 receptor
(IGF-1R). Currently, we are looking at signaling effects of these
antibodies on PCa cells at different stages of the disease, studying
what effects these antibodies might have on the activation of certain
proteins in PCa cells. By performing western blots on protein
extractions from LNCAP, C4-2, and PC3 cells we were able to see changes
in the activation levels of certain proteins caused by treatments with
IGF-1 and the antibodies 1H7 and 3B7. Cells treated with 1H7 and 3B7
expressed higher levels of activated Akt, and lower activation levels
when treated with IGF-1. Phosphorylated tyrosine residues on IRS-2 were
only seen in cells treated with 1H7 and 3B7 whether or not IGF-1 was
added. Phosphorylated serine residues on IRS-2 varied greatly in all
cell lines. Treatment with 1H7 and 3B7 appears to alter molecular
signaling through the IGF-1R.
|
 Investigation of the Presence of Purinergic Receptors in Cancer Cell Lines Julia Greenfield, Christine Maguire, Robert A. Sikes and Kenneth L. van Golen Lincoln University Once thought to be only
involved in the metabolic process, knowledge of the complete function
of the ATP molecule has expend and with it the concept of purine
nucleotides and nucleosides as extra cellular messengers. These
messenger molecules constitute what is known as the purinergic
signaling system, which plays a role in the signaling of many cellular
functions from neurotransmission to cell
differentiation[1]. Previous literature has shown that ATP
signaling mediated by P2 receptors has anticancer effects.
However, other studies have demonstrated a role of purinergic signaling
in cancer progression leading to metastasis. We hypothesize that
certain purinergic molecules are present on prostate cancer cells and
dictate their ability to metastasize. Previous work in our laboratory
has shown a distinct pattern of purinergic receptor expression on the
mRNA level in the LNCaP series of isogenic cell lines. In
addition, these cells produce and release ATP into the extracellular
environment. To illuminate which of the eight P2 receptor are expressed
on the protein level, immunoblots were preformed, selecting for a
number of the P2Y receptors. Currently we have demonstrated
expression of the P2Y1, P2Y6, and P2Y14 receptors which have the
strongest signal. Expression of the remaining receptors need to
be determined.
|
|
Study
of unique microRNA expression signatures in normal colonic epithelium
and colon cancers
Sepehr Sedigh Haghighat, Greg Gonye1, Tao Zhang, and Bruce Boman Department of Biological Sciences, University of Delaware and 1Thomas Jefferson Medical University Dysregulation of crypt
cell proliferation and differentiation has been implicated in the
development of colorectal cancer (CRC). As part of our investigation of
regulatory factors involved in the stem cell origin of CRC, we
investigated the role that microRNAs (miRNAs) might play in colon
carcinogenesis. miRNAs are RNAs 20-24 nucleotides long that were
recently shown to modulate many cellular signaling pathways through
post-transcriptional regulation of messenger RNA (mRNA) levels and thus
protein synthesis. Our previous micorarray analysis (368 gene chip)
showed increased expression of 37 miRNAs in CRCs compared to normal
colonic epithelium. Here we further evaluated miRNA expression in CRC
versus purified colonic epithelium by quantitative PCR (QPCR). Total
RNA was immediately isolated from tissue by the TrizolTM method. RNA
was transcribed into first-strand complementary DNA (cDNA) through
reverse transcription (RT). cDNA was then amplified through QPCR.
Statistical analysis and plots of expression data for miRNAs were done
to find miRNA genes that are differentially expressed. We found, using
QPCR, four miRNAs to be differentially expressed in CRCs versus normal
colonic epithelium including mir-25 and mir-198. Identification of
miRNAs specifically expressed in normal colonic crypts and changes in
their expression in CRCs will provide important information to help
understand mechanisms of colon tumorigenesis.
|
 Testing natural compound extracts for anti-cancer properties Jeneice Hamilton and Kenneth VanGolen Department of Biological Sciences Five natural compound plant
extracts (spices) that are suspected to have anti-cancer properties
were obtained from India. The purpose of this experiment is to provide
scientific evidence to confirm whether or not the spices possess such
properties. Each of the compounds, namely: Zingiba officinale, Tinospora,
Ccucuma Longa, Piper Betel and Piner Longum, was tested on human PC-3
cells. A thousand cells were placed in each well of a 96 well plate.
The following concentrations (in microliters) of spices dissolved in
DMSO was added to the cells: 1:10, 1:50, 1:100, 1:250, 1:500, 1:1000
and 1:2500. Controls were 1:500 and 1:1000. Readings were taken at
different time points, that is, every day for four consecutive days and
the final reading was taken on the seventh day. At the end of each time
point, cells were treated with MTT and incubated for two hours. A MTT
assay was then ran and the data was collected and graphically presented
showing absorbance against time. Based on these data, it was
observed that higher concentrations of each spice adversely affected
the cells and the effects are mostly noticeable at a later time point.
Compound Zingiba officinale
effect on cells is only noticeable by day four however, the change was
not significant. Compound Tinospora effect on the cells was
barely noticeable on day three, but fell on day four and increased
significantly on day seven at the four highest concentrations.
Compounds Piper Betel and Piner Longum showed quite a significant
effect on cells by day four. Compound Ccucuma Longa is still undergoing
evaluation and the final readings for the last four compounds are set
for the next 24 hours. It is however predicted that absorbance of cells
at the higher concentrations will be much less than at the lower
concentrations. This implies that the extracts are either inhibiting
the growth of the cells or killing them.
|
 The Role of Extracellular Matrix on ATP release by Prostate Cancer Cells in Bone Metastasis Dominick Harrison1, Randall Duncan, Robert Sikes, Mary Boggs, and Christine Maguire 1Delaware State University The Effect of Extracellular
Matrix on ATP release by Prostate Cancer Cells in Bone Metastasis
/ / Prostate Cancer is one of the most common types of cancer
found in men affecting one in six in the United States, but it is the
metastasis of this cancer to bone and pain associated with this
metastasis that dramatically impacts the quality of life of the
patient. Unfortunately, the mechanism of how cancer cells metastasize
is unclear. Recent studies have suggested that tumor cells produce
abundant ATP and our lab postulates that the release of ATP from these
cells, and the subsequent binding to purinergic receptors regulates
metastasis. As the cancer cells metastasizes, it encounters and binds
to different types of extracellular matrix (ECM) proteins that we
believe will increase the release of ATP and enhance metastasis of the
cancer. To test this hypothesis, we used the LNCaP (Lymph Node
Carcinoma of the Prostate) progression model of Prostate Cancer. This
model consists of four cell lines at various stages of prostate cancer
progression from relatively benign (LNCap) to highly aggressive and
metastatic to bone (C4-2B). In this project, I grew C4, C4-2, C4-2B
cells on different ECM proteins and determined if the release of ATP
from these cells was dependent on the attachment of these cells to the
ECM. Cells were grown on either tissue culture plastic, collagen I or
neural ECM proteins produced by primary culture of Schwann cells. Basal
release of ATP was measured by removing an aliquot of medium from the
cells grown on these ECM proteins and indirectly measuring ATP using
the luciferin-luciferase assay. My lab has shown that ATP release is
regulated by extracellular calcium entry, so we added either ionomycin
(calcium ionophore) or high KCL (depolarization to active voltage
sensitive calcium channels) to the medium to determine if cells grown
on this different ECM’s had different amounts of ATP available for
release. Department of Biological Sciences, Funded by Department of
Defense
|
 Mathematical Modeling of Colon Stem Cell Dynamics Jennifer M. Hoffman, Bruce M. Boman1, and Gilberto Schleiniger2 1Department of Biological Sciences at the University of Delaware, Center for Translational Cancer Research, Helen F Graham Cancer Center , 2Department of Mathematical Sciences, University of Delaware Previous research has proven
that the overpopulation of colonic stem cells is the ultimate driving
mechanism in tumor growth, which may result in colorectal
cancer. For this reason, a mathematical model is necessary
in order to examine how altered colonic epithelial dynamics give rise
to colonic stem cell overpopulation. A reaction-diffusion model
consisting of a system of partial differential equations were created
to replicate the normal proliferation, differentiation, and diffusion
of the stem cells in the colonic crypt. This model was then coded
and solved using Matlab. Also, this model can be used to explore
how parameter changes can simulate changes in normal tissue and to fit
biological data for premalignant and malignant colon tissues.
Therefore, in the future, these new model parameters can be determined
and the differences between the normal tissue model parameters and the
perturbed tissue model parameters may provide an explanation for how
altered epithelial dynamics lead to colon cancer stem cell populations
that drive tumor growth. This project was funded by the
University of Delaware’s Undergraduate Research Program.
|
 The Presence of JAM-A in Sperm Proves Essential for Normal Sperm Motility: Progressing from Mice to Humans Jean Huynh, Rolands Aravindan, and Patricia A. Martin-DeLeon Department of Biological Sciences There are a number of factors
that lead to male infertility, and among the most important ones is
sperm motility. Junctional Adhesion Molecule - A (JAM-A) has been
recently shown to be a protein that is essential for normal sperm
motility. In mouse sperm, it localizes within the plasma membrane
overlying the head, the midpiece, and the proximal principal piece of
the flagellum. Western analysis of human sperm has revealed the
presence of JAM-A, suggesting that it may also play a role in human
sperm motility. Thus, it is important to extend the analysis of JAM-A
to determine its role in human sperm function. This project is designed
to expand our understanding of JAM-A in human reproduction.
Immunocytochemistry (ICC) was used to localize the expression of JAM-A
in human sperm, which is present throughout the tail and head of human
sperm. Western blots revealed the presence of an ~32 kDa JAM-A band in
human sperm proteins. Lastly, Flow cytometry was conducted to confirm
the Western blot data. Once JAM-A has been fully characterized in human
proteins, the next step will be to co-localize proteins associated with
JAM-A. The localization and characterization of JAM-A in human sperm is
expected to increase our understanding of human male infertility and
subertility, and will be a means of laying the groundwork for assisting
a subset of couples with fertility issues.Funded by NIH-COBRE grant
#5P20RR015588-07.
|
|
Global
whereabouts of three recently discovered single-stranded DNA viral genes
M. Hyatt1, J. Clarke2, W. Kress3, R. Marine4, M. Dumas4, S. Jamindar4, S. Polson6, K. E. Wommack4,5,6 1Delaware Technical and Community College, Newark, DE; 2Dept. of Biological Science, Lincoln University, Lincoln University, PA; 3Dept. of Civil and Environmental Engineering, University of Delaware, Newark, DE; 4Dept. of Biological Sciences, University of Delaware, Newark, DE; 5Dept. of Plant and Soil Science, University of Delaware, Newark, DE; 6Delaware Biotechnology Institute, University of Delaware, Newark, DE. Phages are remarkably abundant
and play critical roles in bacterial population dynamics and
biogeochemical cycling. Studying phage genetics in different
environments can provide clues about their ecological importance and
diversity. Predicted open reading frames (ORFs) from two
single-stranded DNA (ssDNA) viral metagenome libraries were clustered
by homology and ORF clusters were ranked by the number of member
ORFs. Primer sets were developed for three ORFs in the top twenty
clusters. Viruses within water samples were concentrated using
tangential flow filtration. Soil viruses were extracted using
sonication, centrifugation, and filtration with potassium citrate as an
eluant. Using PCR, each primer set was tested on seventeen viral
concentrates from various points around the globe including the
Chesapeake Bay from which two of the clusters were derived, Bergen,
Norway, and Santa Barbara, California. When positive results were
obtained, gel bands were extracted and purified, cloned and
sequenced. Genes that were originally discovered in viral
metagenomes from the Chesapeake Bay and Dry Tortugas were present in
other sites in Delaware, but not in Santa Barbara, California or
Bergen, Norway. Study of single-stranded DNA viruses have been
often overlooked when examining the virome of an environment.
This research has shown that it is possible to selectively target
unique ssDNA genes and test for their presence in multiple
environments. Study of these highly abundant and often unknown
genes will help elucidate what kind of impact ssDNA viruses have on
their hosts and potentially give researchers an assay for detection of
specific viruses in new environments. This research was funded by
the National Science Foundation.
|