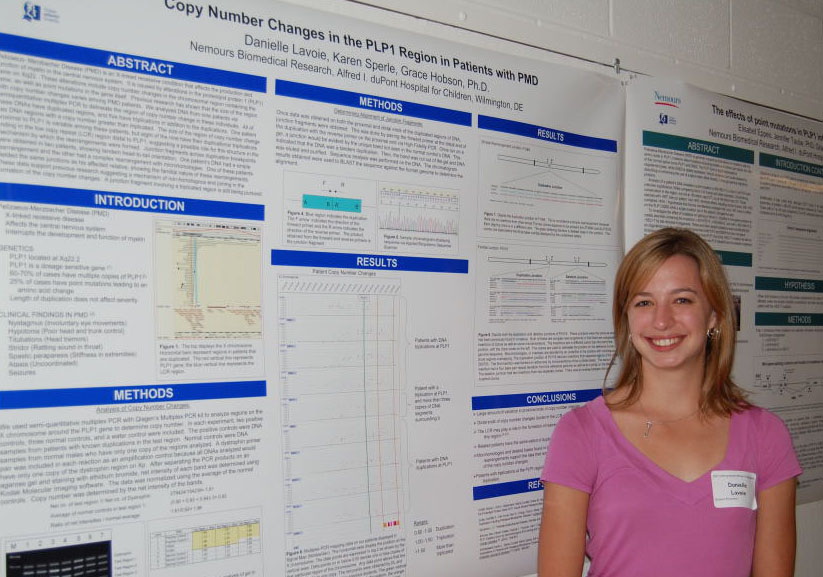
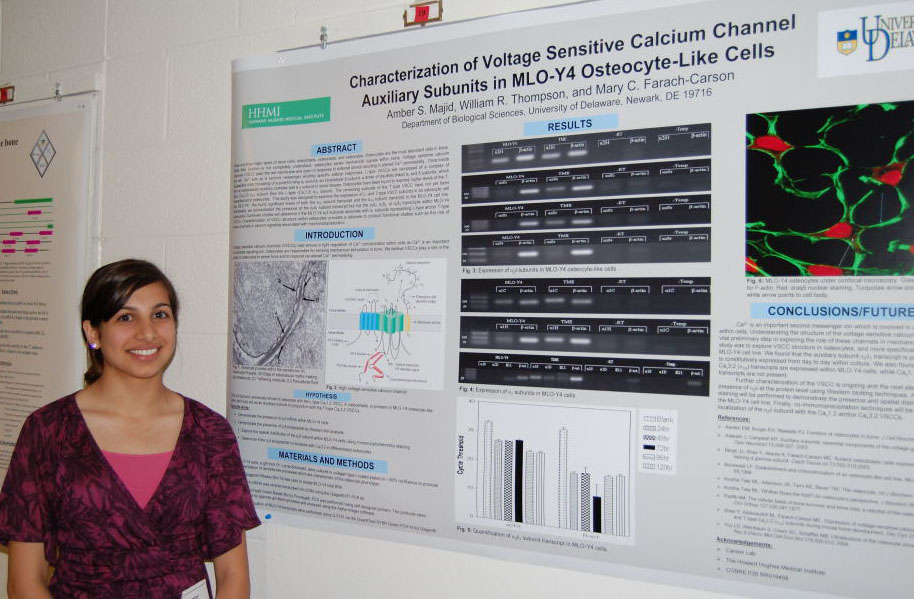
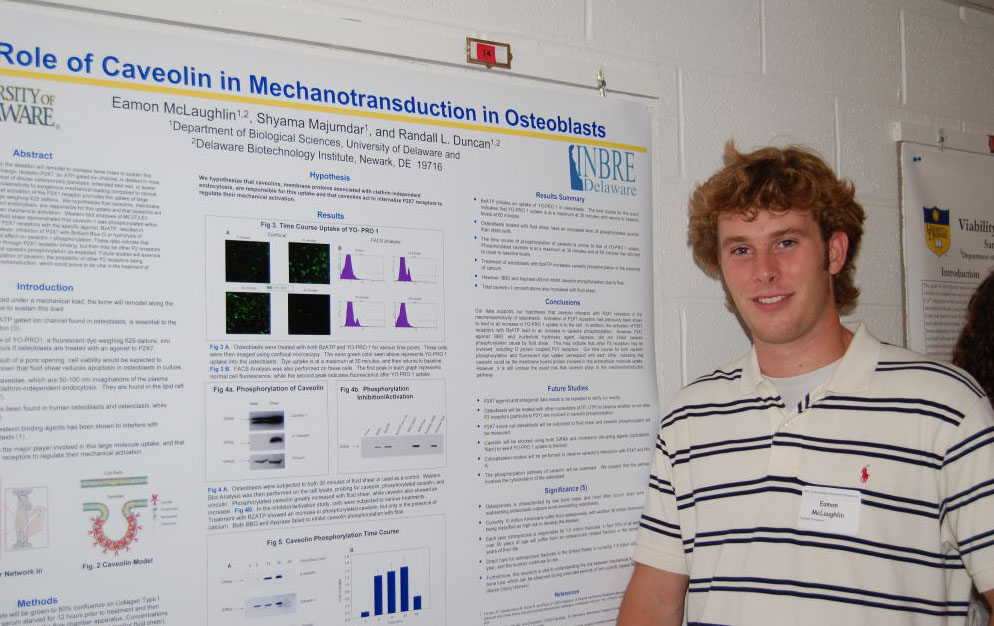
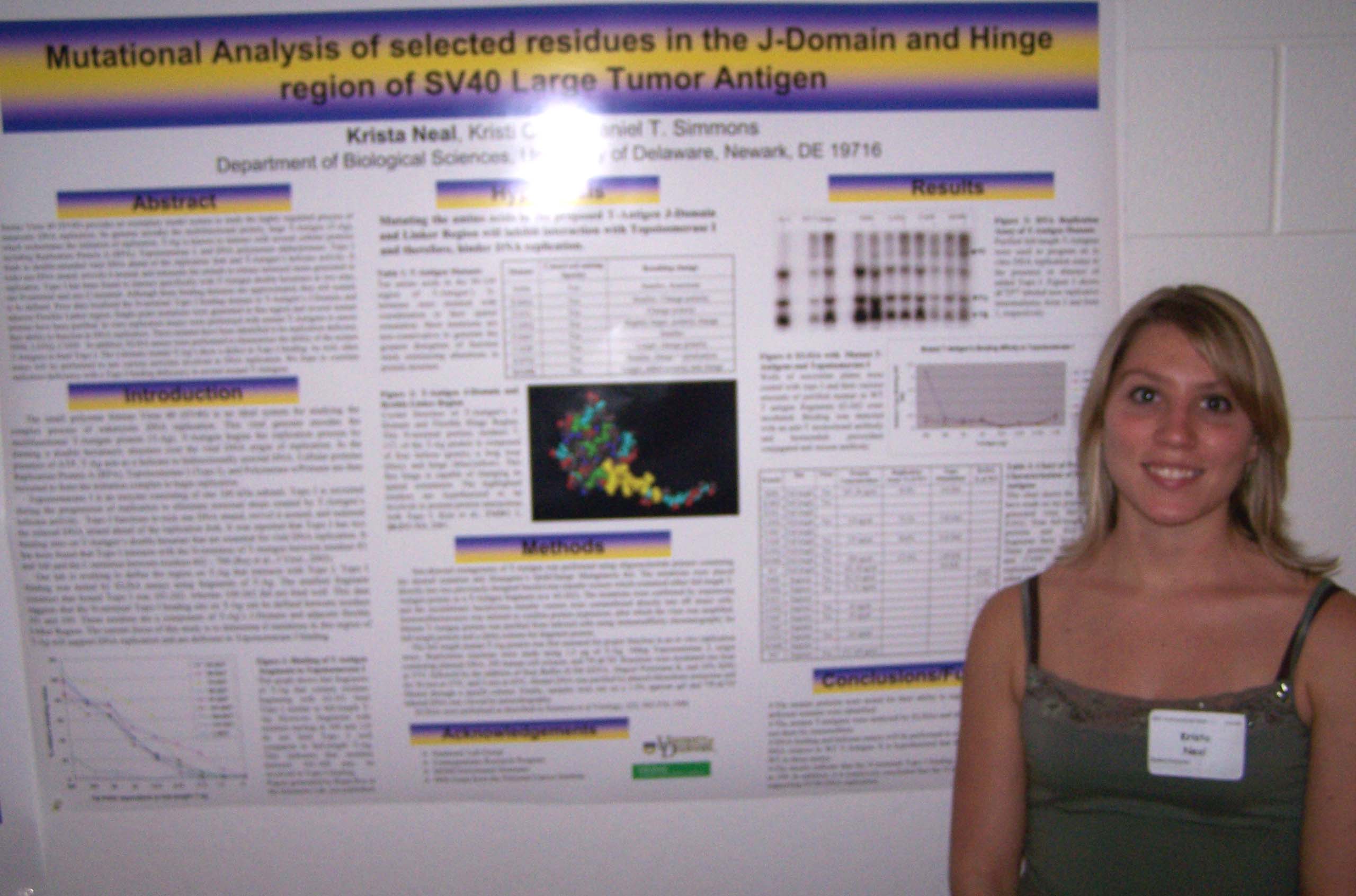
 Regulation of Spot14, a Gene Induced by Thyroid Hormone in Preadipocytes Marysol D. Lavander, Barbara Kwakye Safo, David Usher Department of Biological Sciences Spot14 or Thyroid hormone
responsive protein is a
transcription factor found in lipogenic tissues such as the liver,
brown and white
adipose tissues and lactating mammary glands. In previous studies, it
has been
shown that expression of Spot 14 is regulated by liver X receptor
(LXR), peroxisome proliferator-activated
receptor gamma
(PPARγ) as well as triiodo-L-thyronine (T3). There are two,
long
term goals of this study. One is to identify Spot14 target genes in
adipocytes.
Once a gene has been identified, experiments will be done to locate
Spot14’s
binding sequence. Possible target genes of Spot14 are ATP citrate lyase
(Acly),
malic enzyme supernatant (Mod1), malate dehydrogenase 1 (Mdh1),
acetyl-Coenzyme
A carboxylase alpha (Acaca), pyruvate carboxylase (Pcx), and solute
carrier
family 25 member 1 (Scl25a1). The second goal is to determine regions
of the
Spot14 promoter, which make Spot14 sensitive to cholesterol
concentration. In
this study, primers were designed to identify the proximal promoter
region of
Spot14, a region 2kb upstream of the 5’ end of the gene. Once
identified this
proximal promoter will be inserted into the expression vector pGL4
Luciferase
Reporter from Promega. Primer selection was done by utilizing the
Primer3
website to customize the forward and reverse primers for a 2kb product.
The reverse
primer region was tested along with the HindIII linker that would be
attached
to the Luciferase gene on the vector by using the NCBI Open Reading
Frame (ORF)
Finder. Once it was established that the primers worked, we ran RT-PCR
on DNA from
isolated undifferentiated fibroblast cell line 3T3-L1. Funded by
the Science and Engineering Program as a Life Science Scholar. |