DNA sequencing
UD, DBI use advanced technology in genomic sequencing
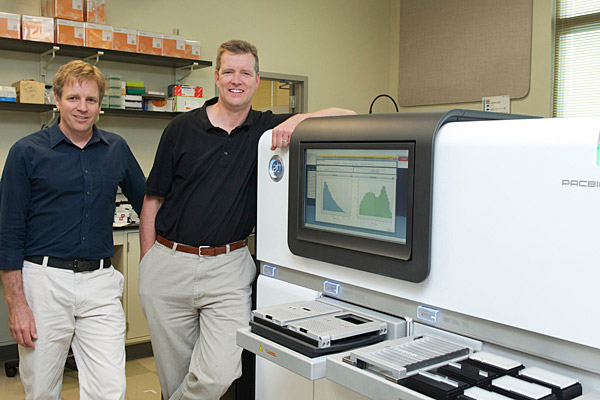
2:48 p.m., May 22, 2012--The Delaware Biotechnology Institute (DBI) at the University of Delaware has been equipped with a state-of-the-art Pacific Biosciences RS DNA sequencing machine to help researchers obtain genetic information, making UD one of the few universities in the country equipped with the advanced device. In fact, at the time of its installation, UD was the 25th site in the world to house such a machine.
The machine came to UD and DBI thanks to a $744,538 National Science Foundation (NSF) award and is stationed in the UD Sequencing and Genotyping Center, which is run by Bruce Kingham and provides genomic technology services for University research groups as well as outside investigators.
Research Stories
Chronic wounds
Prof. Heck's legacy
UD is in a unique spot with the machine because, as K. Eric Wommack, professor of environmental microbiology in the Department of Plant and Soil Sciences, explained, “Most of the places that have the Pacific Biosciences machine are large sequencing centers that sequence the human genome through massive amounts of sequencing, or they’re government labs. I believe we are one of the few research Universities without a medical school that has one of these instruments.”
Because of this, the sequencing machine will be used in unique ways, branching out into areas such as environmental and agricultural studies, with Wommack being one of the first UD professors to use the machine for research purposes.
Wommack was awarded a $200,000 NSF EAGER Collaborative Research grant focusing on exploratory application of single-molecule real time (SMRT) DNA sequencing in microbial ecology research.
The grant will focus on three areas of research. The first is in the area of single cell genomics, in which Wommack and his collaborator at the Bigelow Marine Laboratory in Maine will pull a single bacterial cell out of an environmental sample and sequence its genome.
The second has to do with characterizing the composition of a microbial community using a single gene, with the Pacific Biosciences machine enabling Wommack to look at changes in the sequence of a single gene in order to infer information about the species of microbes making up the microbial community as a whole.
The third application is what he calls “shotgun meta-genomics of viral communities,” which is the heart of his research. Explaining further, Wommack said, “The reason it is called ‘shotgun’ is that we literally are randomly sampling sequences from viruses in the environment.”
Piecing together genetic puzzles
Wommack said a problem with obtaining genetic sequence information is that the process takes a long time when in fact it needs to be as fast and accurate as weather predictions. “What if we could only measure temperature once a week? We really wouldn’t know a lot about how the climate and how weather works,” he said. “But of course, we can measure temperature in incredibly small scales of time. That’s where we need to get in the use of genetic data and environmental science and the Pacific Biosciences instrument gets us a little bit closer to that.”
The machines currently utilized at the center -- while they each serve their own function -- are slower than the new model, which Wommack said, “Allows you to run a lot of samples quickly.” They also provide shorter snippets of genetic sequence.
The current high-throughput DNA sequencers read anywhere from an average of 50 base pairs -- that is, DNA “letters” -- to 400 base pairs, while the Pacific Biosciences instrument can read anywhere from 1,000-12,000 base pairs.
Kingham said of the longer sequences, “Many times when you’re studying genomics, the length of the DNA sequence is everything, so the longer the DNA sequence read for each sample, the better.”
With the shorter sequences, Wommack explained, researchers have to take the little snippets and match them together, like piecing together a jigsaw puzzle. He compared putting together data on the old machines to solving a 1 million-piece puzzle, while putting together data on the new machine is like solving a 100-piece puzzle.
Kingham explained that another big advantage of the Pacific Biosciences machine is that researchers are able to monitor a single DNA molecule as it’s being replicated, as opposed to the other machines in the lab that rely on making copies of the DNA before it can be analyzed. “If you make a copy of something and then you make a copy of that copy, and then you make a copy of the copy of the copy, ultimately what you end up with is likely going to look somewhat different than what you started with,” said Kingham. “The same thing can happen when you copy DNA.”
Kingham also said that he encouraged “anybody that’s involved in life sciences research to consider using that Pacific Biosciences sequencer because it’s so new, there’s so little known about how it can be utilized, and through the ability to analyze a single molecule of DNA, it has the possibility to change the face of genomics research.”
Without distinguished faculty at UD involved in genomics research, Kingham said, it may not have been possible to have obtained the machine. “This is a good representation of the prestige of genomics research at the University of Delaware,” he said. “Many of the scientists are internationally renowned in genomics research. Blake Meyers, Pam Green, Eric Wommack, Carl Schmidt, and others -- these faculty members are internationally renowned in their respective fields of study.”
Kingham went on to say that the machine is “absolutely the cutting edge of DNA sequencing technologies, and UD is really fortunate to have it.”
About the UD Sequencing and Genotyping Center
The UD Sequencing and Genotyping Center is equipped with three DNA sequencing instruments -- the Pacific Biosciences instrument, an Applied Biosystems Sanger sequencer, and the Illumina HiSeq 2000 genome sequencer.
While Kingham said he is thrilled about the new Pacific Biosciences machine, he is also quick to point out that each of the sequencers has its own relevant application area.
The Sanger machine, explained Kingham, “has been around for 40 years, and will probably be around for another 40 years. The advantage to that technology is it produces very high quality data. The disadvantage to it is that it can be very expensive to generate that data.”
The Illumina instrument, Kingham said, generates a large amount of data very inexpensively, but he added that the data might not be of as high a quality as that produced by the Sanger machine.
As for the Pacific Biosciences sequencer, the long length of the sequencing read, coupled with the ability to analyze a single molecule of DNA, cannot be matched by the other instruments.
For more information on the UD Sequencing and Genotyping Center visit the website.
Article by Adam Thomas
Photo by Danielle Quigley








