
|
De Novo Design
of Lanthanide Fingers Christopher
W. amEnde, Mao Ye, Neal. J. Zondlo, and Douglass F. Taber;
Department of Chemistry & Biochemistry |
Ordered alphabetically
by student's last name

|
De Novo Design
of Lanthanide Fingers Christopher
W. amEnde, Mao Ye, Neal. J. Zondlo, and Douglass F. Taber;
Department of Chemistry & Biochemistry |
|
The Photokinetic Analysis
of PRODAN and LAURDAN in Lipid Bilayers:
A Time-Dependent Fluorescence Study Vincent Asiago1, Joanna Lin2, Brad Rowe2, and Sharon Neal2 1Lincoln University, Oxford, PA, 2Department of Chemistry and Biochemistry, University of Delaware This research was funded by National Science Foundation. |
|
Conformational
Effects of Tau and cJun Phosphorylation
Agata A. Bielska, Krista M. Thomas, and Neal, J. Zondlo, Department of Chemistry and Biochemistry This research was funded by grants from the American Heart Association, University of Delaware Research Foundation, and Howard Hughes Medical Institute. |
|
The Importance of Threonine
93 and Serine 94 in Adenylosuccinate Lyase
Meghan Cashman, Mark Segall, and Roberta F. Colman; Department of Chemistry and Biochemistry Funding for this project provided by HHMI. |
|
Characterization of
the ThiI-Persulfide Intermediate formed during 4-Thiouridine Biosynthesis
Glenn D. Christman, Chapman M. Wright, Eugene G. Mueller, Department of Chemistry and Biochemistry Two enzymes, ThiI and IscS, are involved in the modification of uridine to 4-thiouridine in some prokaryotic tRNAs. IscS removes a sulfur atom from free cysteine and transfers it to ThiI, where a persulfide is likely formed at cysteine-456 prior to ThiI effecting the tRNA modification. A fluorescent labeling reagent, I-AEDANS, along with MALDI mass spectrometry and SDS-PAGE, have been used to provide additional evidence for the formation of a persulfide on cysteine-456 of ThiI. Sulfur in a cysteine residue, whether thiol or persulfide, will react with I-AEDANS to displace iodide and form a carbon-sulfur bond. Once the fluorescent label is attached, the protein becomes readily detectable under UV light. Reduction is expected to release the label attached to a persulfide but not a thiol group. In a non-reducing environment, both ThiI and IscS appeared in SDS-PAGE gels when viewed under UV light. When treated with a reducing agent, such as DTT or -mercaptoethanol, the fluorescence of ThiI diminishes greatly, but the IscS band remains very bright. In addition, the tryptic fragment of ThiI containing cysteine-456 connected to the fluorescent label was detected using MALDI mass spectroscopy. The observed mass included one “extra” sulfur, indicating that a persulfide, not a thiol group was modified by I-AEDANS. As expected, this fragment disappeared when the trypsin digest was treated with DTT. Supported in part by the HHMI Program. |

|
AFM Analysis of Patterned
Surfaces
Heather Egolf-Fox, Raphael Yoo, Zhanping Zhang, and Thomas P. Beebe, Jr. Department of Chemistry & Biochemistry Neurons of the central nervous system (CNS) do not regenerate well when severed. Patients with injuries to the CNS would benefit from the discovery of a method for promoting the regeneration of severed nerves after injury. In order to better understand the outgrowth of young neurites under carefully controlled conditions, surfaces were patterned on the micron scale with specific chemical and biological cues using materials affecting neuron growth, and later characterized using state-of-the-art surface characterization tools. Microcontact printing (µCP) was used to produce micron-sized polymer features via an elastomeric stamp. The stamp was coated with a comb polymer known to inhibit neuron outgrowth, and then the pattern was transferred to a polystyrene surface using two variations of µCP. Atomic force microscopy (AFM) was then used to analyze the patterned surfaces. The patterns resembled well trimmed hedges on the micron scale. Increasing the concentration of the comb polymer solution, as well as increasing the pressure used to transfer the comb polymer pattern onto the surface, resulted in greater feature thickness, or hedge height, as expected. Furthermore, the apparent feature thickness increased when AFM analysis was performed in water rather than in air. These results will be compared and discussed. This work is part of a larger project supported by NIH grants (NS43928 and EB00463). |
|
Mammalian Sulfhydryl
Oxidases – a Partial Purification
Olajumoke Fadugba and Colin Thorpe, Department of Chemistry and Biochemistry 
Abstract available at poster session Supported by the HHMI Undergraduate Science Education Program |
|
Evaluation by Site-Directed
Mutagenesis of the Role of Asp-206 and Asp-234 in Metal Binding for Human
NAD-Dependent Dehydrogenase
Mammalian NAD-isocitrate dehydrogenase
(IDH) is activated by ADP and is composed of 3 distinct subunits (2 alpha:1
beta:1 gamma). Sequence comparisons with pig and E. coli NADP
enzymes yield alignment of alpha subunit Asp-206 and Asp-234 with
amino acid residues of these NADP enzymes found in their metal binding site.
We now use a plasmid harboring human cDNA which encodes alpha, beta and
gamma subunits [Huh., JBC, 274, 36866 (1999), Soundar, S. et al., JBC, 2003,278:
52146] to express the enzymes in E. coli. Wild type and mutant
enzymes were purified to homogeneity yielding enzyme with all 3 subunits.
Specific activities of 21.7, 9.0 and 0.19 moles NADH/min/mg were
measured, respectively, for WT, D206N and D234C. Previous mutation of Asp-234
to asparagine gave no activity in the mutant showing that Asp-234 is essential
for activity. D206N exhibited activity nearly half of WT indicating Asp206
is not required for catalysis. Mutation of Asp-234 to a cysteine mutant
yielded Isocitrate and Mn-Km values of 3.4mM and 0.46mM respectively which
are similar to those of WT. Kinetic parameters for isocitrate and
Mn binding for D206N were also similar to those of the WT. Addition of 1mM
ADP decreased the Km-isocitrate for D206N & D234C, similar to that of
WT, indicating Asp-206 & Asp-234 are not involved in ADP activation.
The lower Km-Cd of D234C as compared to WT shows Cys-234 has a better affinity
for Cd. The pH-Vmax profiles for D206N is the same as WT, whereas D234C exhibited
the lower pKa of 5.1, consistent with the pKa representing ionization of
the metal-bound hydroxyl isocitrate. These results indicate that D206 is
not important but D234 is essential for catalysis and metal binding. Supported by the HHMI Program. |
|
Expression of G-Protein
(Gαs) in E. coli
Julie Friedman1, Emily McCusker, and Clifford R. Robinson1, 2, 3 1Department of Chemistry and Biochemistry, 2Delaware Biotechnology Institute, 3Department of Chemical Engineering G -protein coupled receptors (GPCR) are integral membrane proteins of significant pharmacological importance due to their central role in transmembrane signal transduction. Signals are transmitted through the GPCRs to heterotrimeric G-proteins upon external stimuli. All cell types express a variety of GPCRs that are coupled to only a limited number of G-proteins; therefore, both the receptor and the G-protein must discriminate among several signaling partners. This specificity is asymmetric, in that a given receptor typically couples to just one G-protein, while each G-protein may couple with dozens or hundred of receptors. To uncover how these GPCR systems have the ability to achieve discrimination, an array of representative members is needed of the GPCRs subclasses and the G-alpha proteins. Gαs proteins, which are the primary signaling component of the heterotrimer, need to be expressed and purified. To initiate these studies we expressed the human Gαs subunit in E. coli. In preliminary experiments we found that the protein is expressed at low levels. We systematically varied expressed conditions and achieved highest yield of protein using the pET15 vector, with a four hour induction using 1 mM IPTG at 37 °C. We are currently purifying the Gαs protein. Funded by a Life Sciences Scholarship. |
|
Study of Trans-Membrane
Helices Five and Six in the Adenosine A2a Receptor
Kellie Machlus1, Jennifer Kreidler1, and Clifford R. Robinson1, 2, 3 1Department of Chemistry and Biochemistry, 2Delaware Biotechnology Institute, 3Department of Chemical Engineering Although much is known about the structure and corresponding function of soluble proteins, the folding mechanisms and pathways of G protein-coupled receptors (GPCRs) are unknown. In my research, the Adenosine A2a Receptor was studied as a model GPCR. Like all GPCRs, A2a contains seven membrane-spanning alpha-helical domains. The goal of my research project was to use recombinant DNA to create a fragment of the A2a receptor that contains trans-membrane helices five and six. Ultimately, we will use these and similar constructs to learn about the assembly pathway and structure of A2a. PCR was used to produce mini-genes for the two-helix fragment, and restriction enzymes were employed to splice these genes into an expression plasmid. E. coli was grown and used to express the mini-genes for the two-helix peptide. We are currently scaling up these E. coli cultures in order to obtain enough peptide to purify. Once this peptide is sufficiently expressed, it will be purified by Ni-Chelate chromatography. We will then characterize the structure of the two-helix fragment, and measure its interactions with other trans-membrane helices from A2a. This project was funded in part by the Howard Hughes Medical Institute Undergraduate Science Education program. |
|
Using
DNA as a Molecular Hard Drive
Mohammad Malik, Sookhee Hwang, Rusty Hamilton, and Junghuei Chen, Department of Chemistry and Biochemistry Computer hard drives have experienced an exponential increase in storage density from the mid 1950s (0.002 Mb/in2) to today’s current technology (4.1 Gb/in2). However, this rate of advancement is being bottlenecked by the very principal by which hard drives work, magnetic recording. The emerging field of nanotechnology is shifting focus from current hardware design towards miniaturization and use of molecular machines. Here, DNA is used as a molecular storage medium which is capable of achieving storage densities far beyond any current hard drive technology (1 Petabyte/L). By employing current techniques in molecular biology and an address book as a model system, the capacity of DNA as a storage medium is examined. Supported in part by the HHMI Program. |
|
Scanning Tunneling Microscope
Tip-Induced Polymerization
of 10, 12-Tricosadiynoic Acid Confined in Molecule Corrals Samuel Mbugua, Shawn P. Sullivan, and Thomas P. Beebe, Jr., Department of Chemistry and Biochemistry In recent years there has been an overwhelming interest in the understanding and production of “molecular electronics” and nanoelectronic components. 10, 12-tricosadiynoic acid (TCDA), a substituted diacetylene molecule, is an excellent candidate as a means to potentially “wire” new nanocircuitry. It has been shown that TCDA will undergo a topochemical polymerization reaction to form conjugated “nanowires” via a Scanning Tunneling Microscope (STM) tip-induced polymerization method. Polymerization almost always occurred in straight lines along molecular row or a single lamella of TCDA, but in rare cases it was observed that some polymerization occurred across (perpendicular) the TCDA lamella, as well as at domain boundaries, such as at the edges of a “molecule corral.” The oligomers which formed perpendicular to the lamella are believed to be the result of the STM tip interacting with the monolayer at the solid-liquid interface, thus pushing the molecular “line” into a new, less likely orientation. It has been determined that the pulse width as well as the amplitude used to initiate polymerization had an effect on the likelihood of forming a polymerized oligomer. This research specifically focused on using a voltage pulse of U = -6 V and τ = 8 ms to start the polymerization reaction. It was determined that out of 124 pulses applied, 72 polymerized oligomers formed, for an efficiency of 58%. Such a result is significant in providing a large statistical data set with which to evaluate many different pulse widths and amplitudes to better understand the mechanism of polymerization. Experiments using a shorter pulse width (τ = 4 ms) are underway and will be reported when a statistically significant number of data points have been collected. This work was part of a larger project funded by the National Science Foundation. |
|
Tim Martin, Yigang He, and Douglass Taber Department of Chemistry and Biochemistry 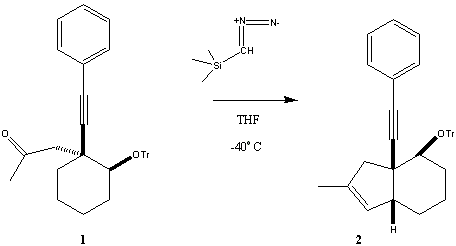
This work is supported by NIH (6M60287). |

Winner of the Sigma Xi Undergraduate Research Award for his oral presentation. |
Role of TM5 in Adenosine
A2A Receptor Dimerization
Matt Roberts, Damien Thévenin and Clifford R. Robinson1, 2, 3 1Department of Chemistry and Biochemistry, 2Delaware Biotechnology Institute, 3Department of Chemical Engineering G-Protein Coupled Receptors (GPCRs), which are integral membrane proteins, mediate a wide variety of signals across the plasma membrane. They play important roles in cell signaling, transport, and metabolism, and are the primary targets of marketed drugs. However, little is known about GPCRs’ molecular mechanism of activation and structure/function relationship. Nonetheless, it is now widely accepted that GPCRs can oligomerize. Oligomerization is believed to be essential in GPCR signaling modulation, but the nature of the interactions is poorly understood. Our goal is to study the interactions involved in the dimerization of an important member of the GPCR superfamily: the adenosine A2A receptor. Among other functions, A2A has been implicated in blood supply regulation, vasodilatation, and in various neural degenerative diseases. Our approach is to study A2A dimerization through the association of peptides corresponding to the receptor’s TM domains. Circular dichroism (CD) spectroscopy, Förster resonance energy transfer (FRET), and gel electrophoresis indicate that TM5 can self-associate to form higher-order oligomeric states. This suggests a possible role for interactions between TM5 domains in the oligomerization of the A2A receptor. Supported in part by the HHMI Undergraduate Science Education Program and an NIH COBRE grant.. |
|
Development of New Cross-Coupling
Reactions
Rebecca Salomon and Neal J. Zondlo, Department of Chemistry and Biochemistry Abstract Available at Poster session. Funding provided by a Howard Hughes Medical Institute grant. |
|
Probes for Molecular
Presentation in Neuron Guidance
Christopher Schaffers, Vinod Tarwade, Zhenzhen Dong, Katherine Drake, John Koh, Joseph Fox, Department of Chemistry and Biochemistry Ephrin-A5 is a repelling neuron guidance cue involved in the formation of retinotopic maps during neurogenesis. Ephrin tyrosine kinase receptors are positioned and can move freely along cell surfaces as the axon stretches and utilizes these receptors in pairs separated by a biologically determined distance. It is the purpose of this multidisciplinary research group to develop methodology to ligate desired proteins such as ephrin receptors to either end of a rigidly linear organic molecule, departmentally coined as The Nanorod, with biologically relevant lengths of between 10 and 100 nanometers. With protein receptors equally spaced, the nanorod will be attached to a thin gold film or glass for analysis and further experimentation on axon outgrowth with varying nanorod lengths and cluster formations. Supported in part by the HHMI Undergraduate Science Education Program. |
|
Enzyme-Product Complex
of Enoyl-CoA Hydratase
Anisha Vora and Brian J. Bahnson, Department of Chemistry & Biochemistry Enoyl-CoA hydratase (ECH) is an enzyme involved in fatty acid metabolism, which catalyzes the syn addition of water across the double bond of alpha, beta-unsaturated CoA thiolesters. An E. coli over expression preparation of the enzyme, which is in an expression plasmid with an IPTG chemical control, was performed to obtain 20 mg of pure ECH per L of cell culture. The expression, preparatory work, and ion exchange chromatography was used to purify the protein according to the experimental procedures developed in the Bahnson lab. The most useful purification step of ECH utilized protein crystallization and recrystallization using ethanol as a precipitant to obtain pure enzyme. The next step will involve growing diffraction quality crystals with the bound product, propionyl-CoA and solving the crystal structure. A true product-complex will give further insight into mechanistic details of this enzyme. Supported by the Science and Engineering Scholars Program and the State of Delaware. |