Instructor: David Kirchman (kirchman@udel.edu)
September 4, 1997
MAST 634
Lecture 1: Overview
Molecular Taxonomy
Classical methods don't work; inadequate
What organism is it?
Microbes only way
Other problems
Stock Identification
Invading species
Aquaculture
General population biology
What
is molecular taxonomy?
Using molecules to identify organisms, specifically the sequence
DNA sequence
Deduced amino acid sequence for protein
Several molecules used; see homework problem
Concentrate here on small sub unit RNA
Review
Prokaryotes = bacteria and archaea
Eukaryotes = everything else
See
table for differences in cellular organization
Brief review of ribosomes
-know
role of ribosome in protein synthesis
p 982
and 989 for details
Complex
structures
several proteins
SS 70 S
21 polypeptides
LS 80 S
49 polypeptides
Miscellaneous information about ribosomes
See Table
40% of ribosome is protein
ribosomal protein 15%
of total protein
protein is 60% of cell mass
ribosomal RNA = rRNA; genes for rRNA = rDNA
rRNA up to 30% of cell mass
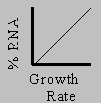 not always true for eukaryotes
not always true for eukaryotes
Remember: 80% of RNA is rRNA
Fundamental problem in Marine Biology and Oceanography: determining
growth rates in the field
Two major ribosome types
|
70S
|
80S
|
| Organisms |
Bacteria
Organelles |
Eukaryotes |
| Small Subunit |
30S
RNA 16S 1542 nt) |
40S
RNA 18S (1874 nt) |
| Large Subunit |
50S
RNA 23S (2904 nt)
5S (120 nt) |
60S
RNA 285(4718 nt)
5.8
5S |
| Sensitivity |
chloramphenicol |
cycloheximide |
Page 1002
in textbook for others
Antibiotics useful experimental tool
plants have different rRNA
Some work on 5S, but mostly 16S and 18S rRNA
Why 16S?
1. All
organisms have it
2. Enough
data, but not too much
5S ~ 120 nt
16S 1542
23 2904
3. Conserved
regions but some variable
all organisms
only Eucarya, bacteria
selected groups of organisms
- gets
fuzzy on whether you can determine species
Why conserved? Determined by 3-D structure.
See picture of SS rRNA in textbook; good test of sequence is to reproduce
structure
Why is it important to have conserved regions?
1. Have to have
variable because that's how you determine what's different
2. Conserved
regions important in finding variable (different) regions
Variable
5' conserved...............................................conserved
3'
What have we learned from 16S?
- not really good
for stock ID
I. Origin of eukaryotic cell
- look at rootless
tree
- chloroplasts similar
to cyanobacteria
same type of P.S.
- mitochondria similar
to heterotrophic
bacteria, i.e. [CH2O] + O2 H2O + CO2
Endosymbiotic Theory
Eukaryotic cell = collection
of prokaryotic cells
organelles
mitochondria = heterobacteria
chloroplast = cyanobacteria
Other evidence
1. Modern day symbiosis
2. Presence of some genes, but not all in organelles see tables
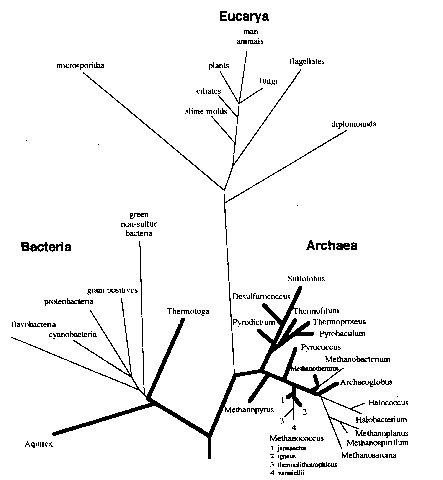
II. Recognition of 3rd Domain: Archaea
Carol Woese
- very different based
on 16S RNA, although they "look" like bacteria
- thought to be ancient
found in extreme environments, at first.
High temperature > ~ 80 C
weird metabolism (more later)
But now found everywhere
Table showing difference between 3 domains
Not so ancient
closer to eukarya?
Controversy pt.
Some evidence for similarity to Eukarya
Archaean
Methanococcus jannaschii
completely sequenced
Hydrothermal vents 2600 m
Grows high pressure
Temperature optimum = 85 C
Methanogen = makes methane
CO2 + H2 CH4 + cell biomass
Unique to archaea
Compare sequence with known genes
- Identify open-reading frames ORF
long stretches of
sequence that can be translated to a protein
without
hitting a stop
DNA AA
sequence
Ribosomal proteins in M. jannaschii
All
++
Archaea-only ++
Bacteria only -
Elongation factors more similar to Eukarya than
bacteria
III. Origin of Life: 3rd major finding from 16S data
Deeply-branched
microbes are Hyperthermophiles
Organisms
grow with optimum temperature 80-110 C
unable to grow <60 C
Record
temperature ~ 160 C ; have to have high pressure for T >100 C
How can
they survive? No simple answer
some have high G-C
G C stronger than A=T
ether linkage in archaea membranes
 Where are they found?
Where are they found?
Hot springs
Yellowstone National Park
Hydrothermal
vents
Very difficult to culture these organisms
In fact, > 99% of all microbes cannot be cultured
How do we determine what species are present?
How do we find 16S rRNA genes = (rDNA) in complex genome?
PCR = polymerase chain reaction
1. 
Known sequence = primers of ~ 20 nt
2. Any other DNA can be present
3. Amplifies = makes many copies of unknown sequence, substantial
above other
DNA
4. Key: heat to high temperature to separate strands. Originally used
heat-labile enzyme
Original DNA polymerase:
Taq: Thermus
aquaticus
Found by Thomas
Brock in Yellowstone
Kary Mullis:
method presented 1985
Nobel Prize 1993
See description in textbook
Can be used
Single organisms
Single cells
Mixed microbial assemblages |

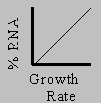 not always true for eukaryotes
not always true for eukaryotes
