| Instructor: David L. Kirchman (kirchman@udel.edu)
September 25, 1997
MAST 634
Lecture 7: Fermentation
Major Points from Last Lecture
Uses of fluorescence
inverse (sort of) relationship between fluorescence and photochemistry
Overview of Metabolism
all lead to central metabolism
Calculating theoretical energy yields
ATP production: substrate - level versus
oxidative ( and photo-) phosphorylations
Fermentation
recycles NAD+
NADH oxidization releases energy as
heat; in oxidative phosphorylation, it drives PMF that in turn drives ATP
synthesis
End products of glycolysis: pyruvates + 2 ATP + 2 NADH; see handout
Without O2: Fermentation oxidizes NADH to NAD+
Ability to deal with anoxia (no O2) ancient trait world
anoxia for first 10a years
oxygen ---> free radicals (e.g. O2-)
and perioxide (H2O2)
Miscellaneous Notes about Lactate and EtOH Fermentation
1. EtOH fermentation
Not a single step reaction
ADH
pyruvate ----> acetaldehyde + CO2 ---->
ethanol
[see page 466 in V&V]
2. Lactate fermentation very common
Lactic acid bacteria do only this
Homofermentation
---> only lactate
heterofermentation
---> lactate +EtOH + CO2
These bacteria need very complex media with many compounds
Trademarks of fermentations
1. Starting organic material partially oxidized
2. Little or no CO2 released; organic
C is released
3. Modest ATP yield, e.g. one or two ATP/reaction
Anaerobes: living without O2
Generally bacteria
Some eukaryotes, but few
Interesting eukaryote: protozoan pathogen,
Giardia
1. Has no mitochondria;
appears to have lost them during evolution
2. ATP produced
by glycolysis and mixed acid fermentation, i.e. several acids are produced.
How do cells and animals normally living with O2
deal with anoxia?
First, consider cells:
Different muscle cells, different levels
of glycolysis, etc (substrate - level phosphorylation enzymes) vs. Respiration
(oxidative phosphorylation)
V&V has bird examples
Let's consider fish examples
Two basic muscle types
1. Red - slow twitch  Sustained E production
Sustained E production
Red color from hemes (Fe) in mitochondria
2. White - fast twitch
short bursts
High amounts of glycolytic and fermentation enzymes in white muscles.
Consider tuna: 40 mph for ~ 10 min with 30 sec between bursts.
High amounts of LDH in tuna white muscle
LDH = lactate dehydrogenase
But when you go into training, you don't increase red muscle ---> enzyme
levels change
Controls on LDH activity
LDH stimulated by anoxic conditions
LDH inhibited by oxic conditions
|
O2
|
pH
|
[pyruvate]
|
creatine -P
|
| oxic |
+
|
~7
|
low
|
high
|
| anoxic |
-
|
low
|
high
|
low
|
what causes low pH in anoxia?
Glucose ---> lactate + 2H+
But not real reason, according to Hochachka and Somero
Real reason: ATP hydrolysis
ATP + H2O ---> ADP + Pi
+ H+
Aerobic respiration re-uses H+
Cells with high LDH activity have ability to withstand low pH (=buffering
capacity)
Deep-sea
fish: float and wait, low LDH activity, low buffering capacity
Warm-bodied
fish: active, high LDH activity, high buffering capacity
Whole animals withstanding anoxia
Important in marine environments --->
sediments
Examples
parasitic helminths
burrowing annelids ("worm")
Intertidal bivalves (at low tide)
Some vertebrates
Goldfish can survive without O2 for several days
Freshwater turtles
Strategies
I. Storage of C: glycogen
important because anaerobic metabolism yields less ATP
Also true in white muscles of aerobic organisms
Examples:
anoxia - tolerant goldfish 1300 mu mol gluc./gm
anoxia - intolerant trout 235 mu mol gluc./gm
Mytilus: 50% of dry wt can be glycogen!
What is glycogen?
See p485 in V&V
1. Alph (1 --> 4) glucose polymer
2. branches 1-6 every 8-12 glucoses
3. occurs as granules intra cellular (very large)
Why glycogen and not fat in muscles?
1. Muscles degrade glycogen faster
2. Fatty - acids (FA) can't be degraded
anaerobically --> needs O2
- O2 not directly involved
- O2 needed for NADH and FADH2 oxidation
 oxidation oxidation
Fan - acetyl CoA + NAD+ + acetyl-CoA ---> Fan-2
- acetyl CoA + NADH
see page 669 in V&V
3. V&V says animals cannot convert fatty acids to glucose (and eventually
to riboses for nucleic acids)
Glucose synthesis: need when animal is "living"
on protein (meat or fasting!)
= gluconeogenesis
phosphorylase
glycogenN + Pi ---------------->
glycogenn-1 + glucose -1-P
G-1-P then enters glycolysis
First of many examples where "starting point"
for glycolysis is not glucose
II. Switch to more efficient fermentation in Mytilus
In Mytilus, for example
|
#ATP's
|
Survival without O2
|
| lactate |
2
|
3 days
|
| propionate |
6
|
9
|
Other important fermentations
In cephalopods: Squid can swim very rapidly uses protein and amino acids
as energy source
specifically: arginine~P (<----- arginine +ATP)
Arg~P ---> ATP + Arg
But what to do with Arg
Arg is very basic ---> osmotic problems
Arg + pyruvate ----------> octopine; octopine is a 9C amino acid
NADH -----> NAD+
octopine is good for osmotic balance
Use of amino acids (actually, proteins that is hydrolyzed to amino acids)
Leu ------------------> Keto isocaproate
 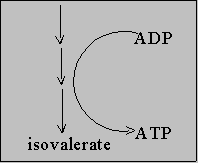
Aerobic degradation
AA ------> alpha - keto acid ---> Kreb
cycle + NH4+
III. Reduce metabolic rate
much better
than switching fermentation
e.g. Decreasing metabolism in Mytilus survival goes
from 3 to 60 days
Effects ATP turnover --but ATP conc. decrease little
|
% Aerobic Rate
|
LT50
|
| Locust |
27 |
72h |
| Mytilus |
5 |
35 d |
| Artemia |
<5 |
>24 months |
Other effects on metabolism (Hand and Hardewig 1996)
Protein synthesis decreases
1. Some evidence that transcription is not affected
2. But genes for some proteins regulated "up regulated", i.e. they
are induced
3. Translation level control?
Protein turnover
both synthesis and degradation of protein
Generally low in prokaryotes: "unwanted" proteins diluted out as cells
continue to divide
Important in eukaryotes
Big decrease in protein degradation
with switch to anaerobic metabolism
Remaining degradation shifts to ATP
independent hydrolysis
How is protein degradation slowed down?
1. Not by directly inhibiting proteases
2. Fewer proteins tagged by ubiquitin;
ubiquitin is a protein. See pages 101-1015
Several ubiquitins
covalently link to doomed protein
In eukaryotes, intracellular hydrolysis of selected proteins carried
out y proteosome, structure similar in size (26S) and complexity as ribosomes.
protein -- ubiquitin +ATP
-------> peptides
proteosome
Half-life of proteins set by structure
PEST proteins
very labile
P Pro
E Glu
S Ser
T Th
applies to prokaryotes too!
|

