
The Zondlo Research Group
Laboratory of Organic Chemistry
and Molecular Design
Current Research
Our research focuses on the design, synthesis and development of small molecules and minimalist polymers with biological activity, the elucidation of fundamental principles of and discovery of effectors of biological interactions, the development of novel, functional proteins, and the development of novel and practical methods of synthesis and enantioselective catalysis.
Our interests center on the general area of functional molecular recognition: the generation of novel molecular structures and architectures which interact specifically with target molecules. Biological targets represent a considerable test of our knowledge of the fundamental principles of molecular recognition. Effective modulation of biological events requires the generation of molecules with both high affinity and high specificity for the desired target. Researchers in my group utilize modern methods of organic synthesis and catalysis, combinatorial synthesis and high-throughput analysis, solid-phase and expression-based peptide and protein synthesis, advanced NMR and computational analysis, molecular biology, and biological assays.
PROTEIN PHOSPHORYLATION AND POST-TRANSLATIONAL MODIFICATIONS
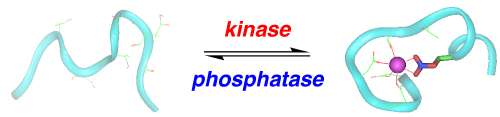 The complexity of humans is dependent on post-translational modifications of proteins. The most common post-translational modification is the phosphorylation of serine, threonine, and tyrosine residues by protein kinases. Protein kinases are tightly regulated, and changes in kinase activity are associated with most human diseases, including cancer, heart disease, and Alzheimer's disease. We are developing approaches to understand how protein phosphorylation changes the structure of proteins toward understanding the mechanisms associating changes in kinase activity with human diseases. In addition, we are developing new tools to understand the changes in kinase activity associated with human disease. We have designed new protein structures, called protein kinase-inducible domains, whose structures are dependent on their phosphorylation state. These designed proteins are under the control of specific protein kinases and are non-fluorescent when not phosphorylated, but highly fluorescent when phosphorylated, and may act as genetically encoded sensors of protein kinase activity.
The complexity of humans is dependent on post-translational modifications of proteins. The most common post-translational modification is the phosphorylation of serine, threonine, and tyrosine residues by protein kinases. Protein kinases are tightly regulated, and changes in kinase activity are associated with most human diseases, including cancer, heart disease, and Alzheimer's disease. We are developing approaches to understand how protein phosphorylation changes the structure of proteins toward understanding the mechanisms associating changes in kinase activity with human diseases. In addition, we are developing new tools to understand the changes in kinase activity associated with human disease. We have designed new protein structures, called protein kinase-inducible domains, whose structures are dependent on their phosphorylation state. These designed proteins are under the control of specific protein kinases and are non-fluorescent when not phosphorylated, but highly fluorescent when phosphorylated, and may act as genetically encoded sensors of protein kinase activity.
SMALL MOLECULE PROTEOMIMETICS
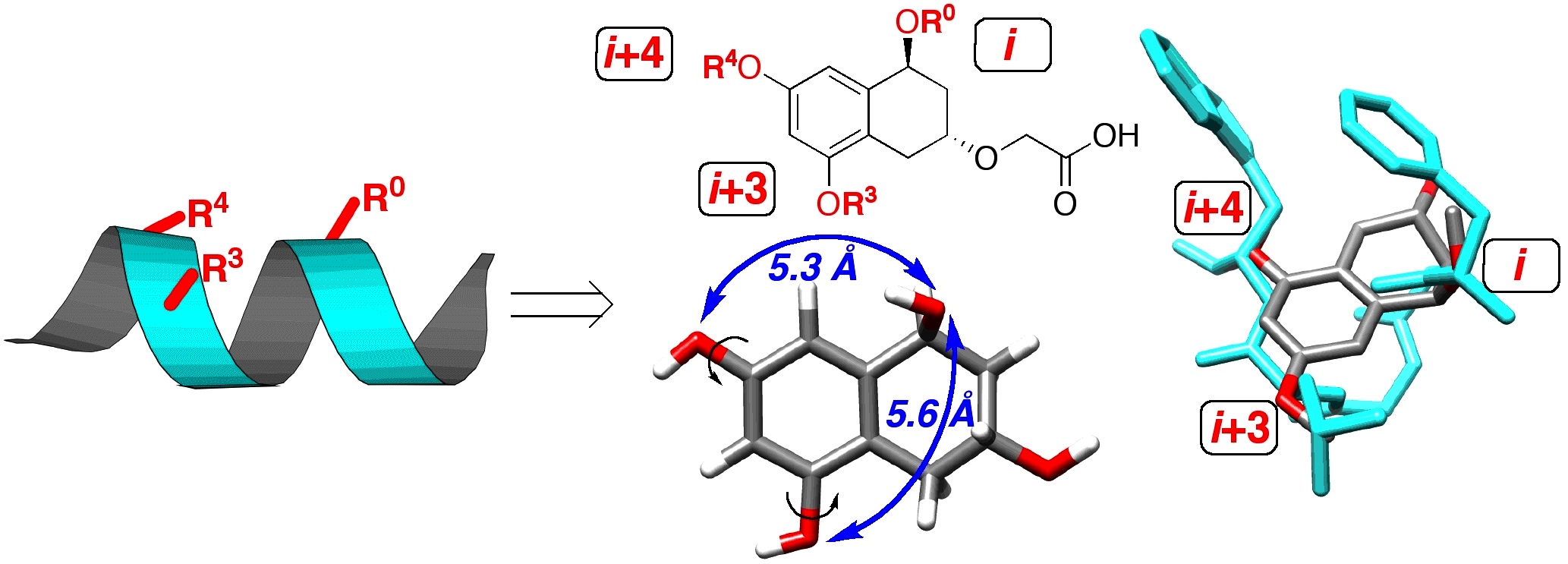
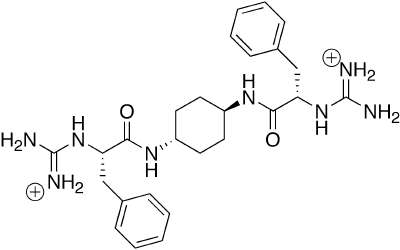 Work in genomics and proteomics is revealing vast numbers of interaction loci within the cellular milieu, and thus vast numbers of potential targets for agonists and antagonists of protein-protein, protein-DNA and protein-RNA interactions. Our focus is the use of molecular design and organic synthesis to develop small molecules which mimic larger biological structures. Our work involves the development of appropriate, readily accessible organic scaffolds, in solution and on solid phase, using modern methods of organic synthesis. To evaluate our scaffolds they are subjected to high-throughput testing for biological activity. Modularity in synthesis allows the combination of multiple structural elements to allow recognition of larger protein surfaces and the synthesis of multifunctional "proteins." The post-genomic era requires novel tools to elucidate the identity, classes and modes of protein-protein interactions in disparate cell types, developmental stages, and intracellular environments, in addition to changes due to age and disease states. The ability to generate small molecule mimics of protein recognition elements permits their use as chemical probes of protein-protein interactions.
Work in genomics and proteomics is revealing vast numbers of interaction loci within the cellular milieu, and thus vast numbers of potential targets for agonists and antagonists of protein-protein, protein-DNA and protein-RNA interactions. Our focus is the use of molecular design and organic synthesis to develop small molecules which mimic larger biological structures. Our work involves the development of appropriate, readily accessible organic scaffolds, in solution and on solid phase, using modern methods of organic synthesis. To evaluate our scaffolds they are subjected to high-throughput testing for biological activity. Modularity in synthesis allows the combination of multiple structural elements to allow recognition of larger protein surfaces and the synthesis of multifunctional "proteins." The post-genomic era requires novel tools to elucidate the identity, classes and modes of protein-protein interactions in disparate cell types, developmental stages, and intracellular environments, in addition to changes due to age and disease states. The ability to generate small molecule mimics of protein recognition elements permits their use as chemical probes of protein-protein interactions.
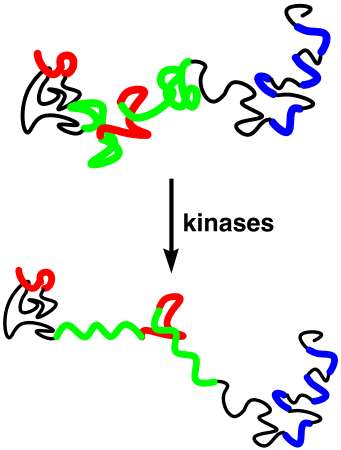
PROTEIN MISFOLDING AND DISEASE
Numerous diseases, including Alzheimer's disease, Parkinson's disease and spongiform encephalopathies (i.e. mad cow disease and its human variants), are characterized by protein misfolding and precipitation which is central to the observed pathology. We are interested in the molecular mechanisms leading from the soluble, monomeric protein to the insoluble, polymeric protein forms. We are particularly interested in understanding the molecular mechanisms of Alzheimer's disease. Alzheimer's disease is characterized by two protein aggreagates in the brain: extracellular plaques and intracellular neurofibrillary tangles (NFTs). The major protein in neurofibrillary tangles is a hyperphosphorylated version of the protein tau. Tau normally stabilizes the elongated structure of neurons by binding to the microtubules. However, hyperphosphorylation of tau, phosphorylation on over 30 residues of tau, results in structural changes in tau that cause the precipitation of tau in neurofibrillary tangles. We are examining the molecular mechanisms by which hyperphosphorylation of tau leads to structural changes, protein aggregation, and neurofibrillary tangle formation. We have found that phosphothreonine induces a particularly ordered conformation.
SYNTHETIC METHODOLOGY: CROSS-COUPLING REACTIONS, STEREOSELECTIVE SYNTHESIS, AND SELECTIVE CATALYSIS
We are interested in developing enhanced proteins via the development of practical approaches to unnatural amino acids to complement the natural 20 amino acids. We have developed approaches to synthesize unnatural proline analogues in a manner to allow control of structure and the introduction of novel functional groups within proteins, including groups for spectroscopic probes and for bioorthogonal chemistry. We have become particularly interested in the amino acid 4-thiophenylalanine, a redox-active analogue of tyrosine with a reduced pKa (pKa 6) and enhanced reactivity. 4-Thiophenylalanine is a versatile probe of sulfur redox chemistry and is a functional hybrid of tyrosine and cysteine. In addition, effective, selective catalysis requires molecular functionality sufficient for catalytic activity, (regio- and stereo-) discrimination in substrate recognition enforced by reproducible transition state geometry, rapid association of substrate and dissociation of product to ensure turnover, and a partially open geometry to allow significant substrate scope. The incorporation of catalytic functionality within a designed structure is an important goal, and provides a critical test of our knowledge of the fundamental principles of molecular folding and catalysis. Due to their inherent chirality and structure, peptides and designed proteins are ideally situated to function as highly effective catalysts, despite limited success to date. Our approach is to design novel peptides cabable of functioning as effective catalysts. A second element to catalyst discovery is the ability to rapidly screen catalyst candidates. To address the scope of combinatorial space, both in terms of catalyst structure and substrate generality, we are developing new methods for high-throughput screening for catalysis. These methods are designed to be applicable not only to the discovery of protein- and peptide-based catalysts, but also toward the discovery of metal-complex-based catalysts and organocatalysts.
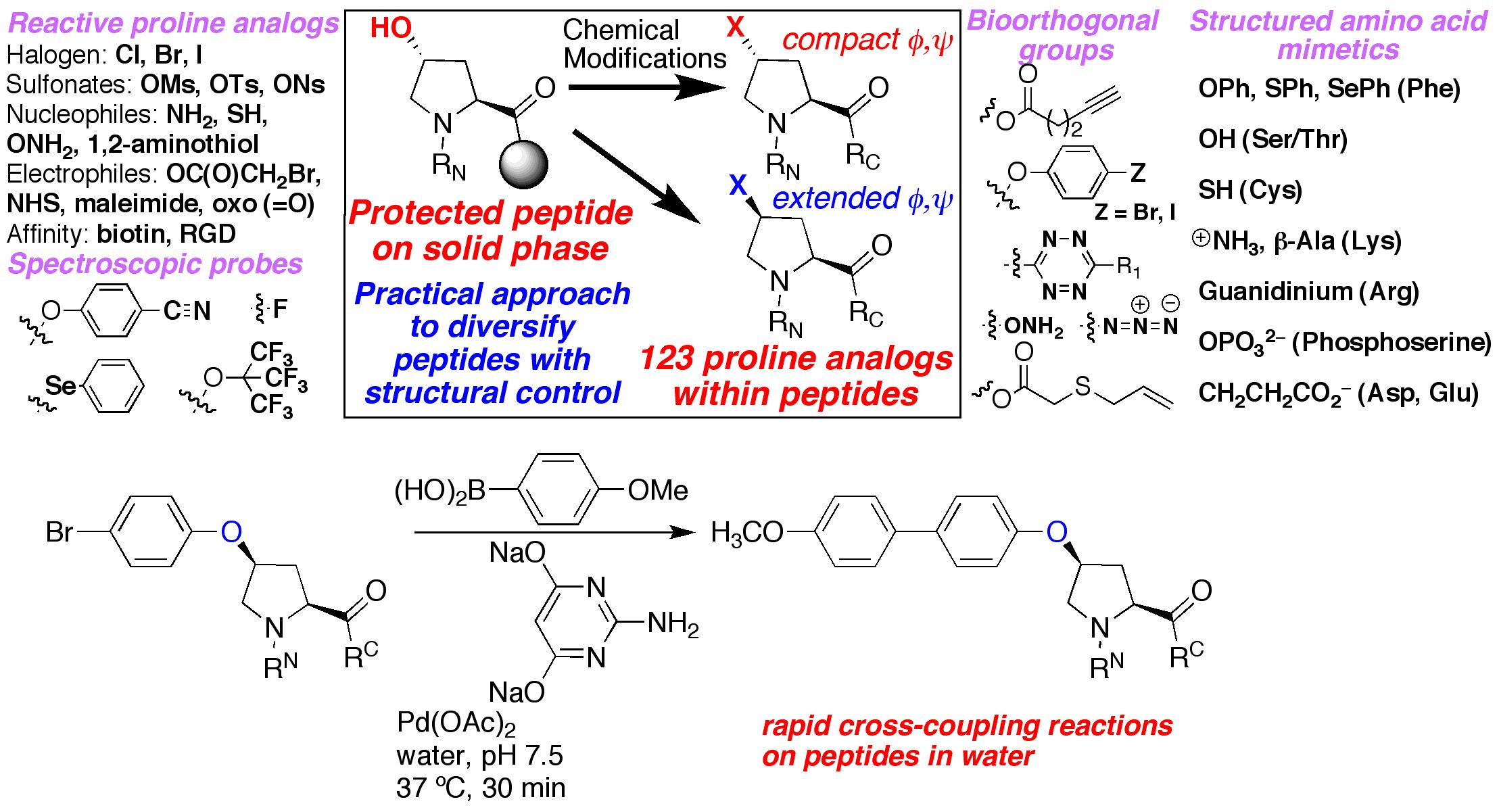
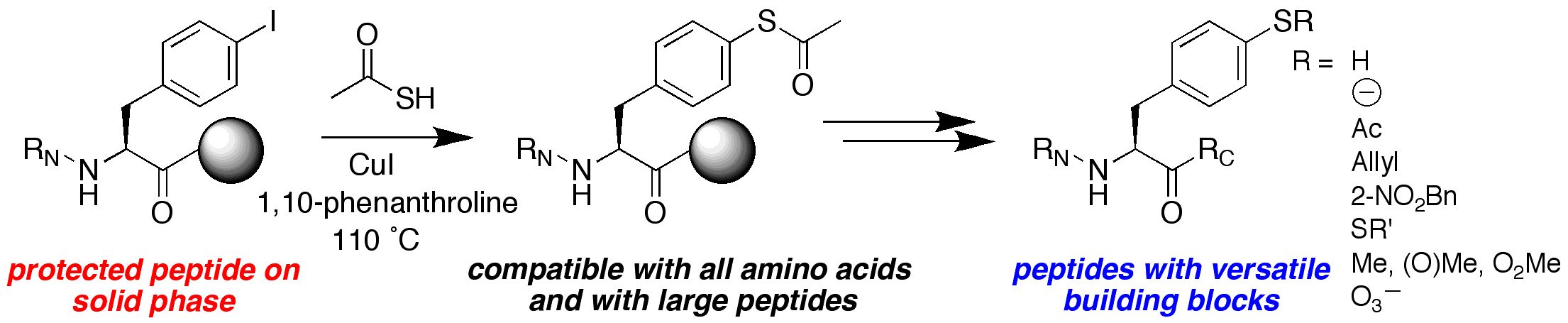
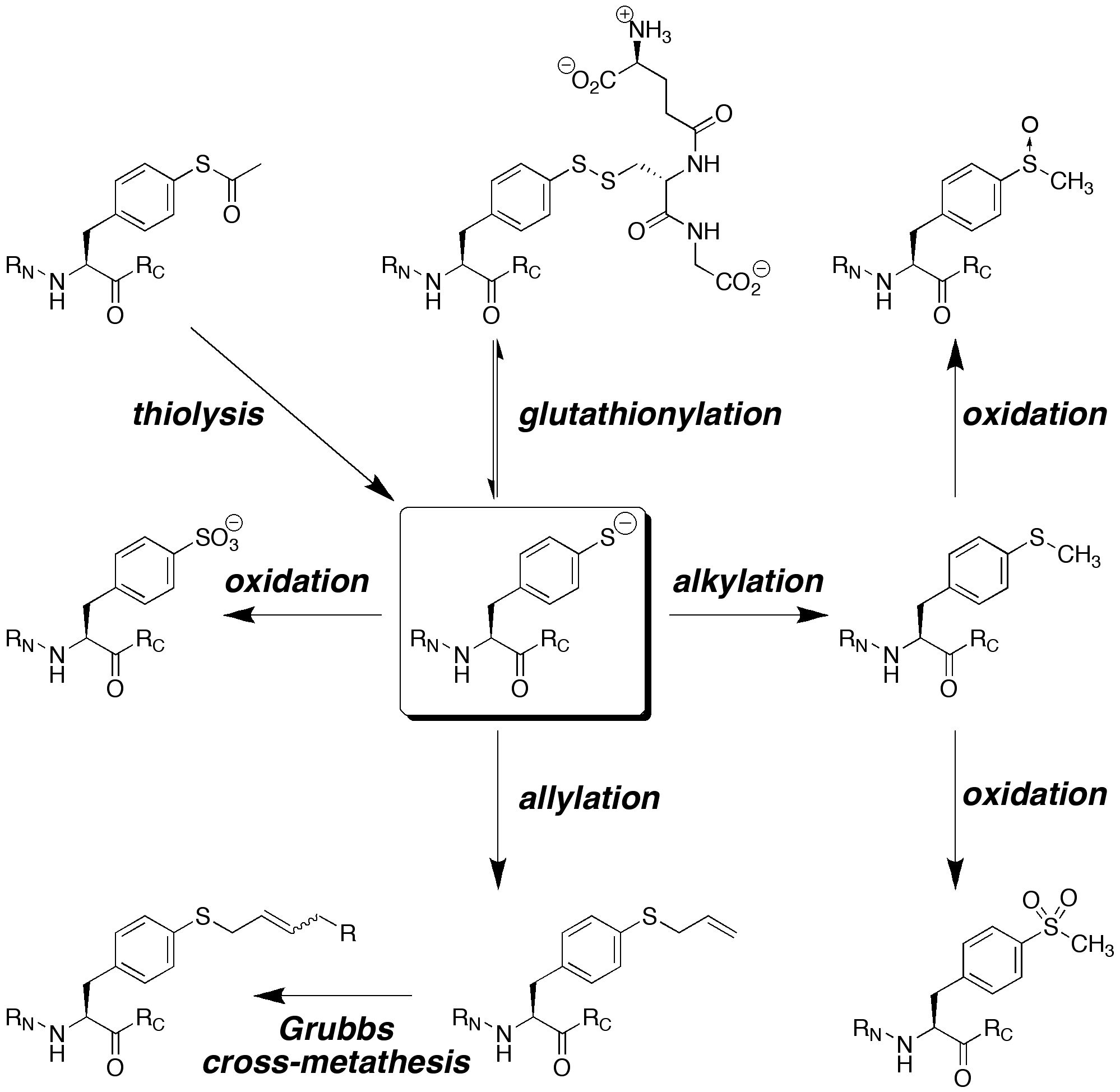
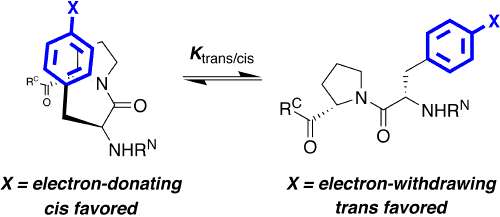
ELECTRONIC AND STEREOELECTRONIC EFFECTS IN PROTEINS AND THE DESIGN OF TUNABLE PROTEINS
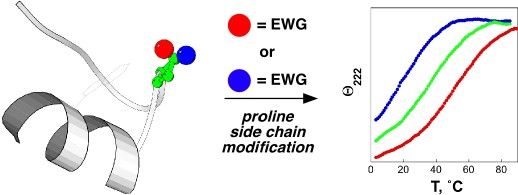
One hallmark of native, functional proteins is the adoption of a stable, highly conformationally restricted ensemble of closely related structures. In peptides, small proteins, and natively disordered proteins, where the hydrophobic effect is reduced, it is difficult to overcome the entropic cost of conformational restriction. Strategies to enable conformational restriction and stabilization of peptides and proteins may be used to interrogate protein structure-function relationships and to develop novel mediators of biological activity. We are developing new approaches to control local conformation via small peptide motifs and practical and readily applied organic synthesis. We are examining two approaches to control protein conformation: controlling main chain conformation via controlling the ring pucker of proline residues, and controlling cis-trans isomerization via tuning of proline and/or aromatic ring electronics. We have demonstrated that the interactions between aromatic residues and proline residues are controlled by aromatic electronics. We are developing approaches to tunably control the structures of peptides and proteins using stereoelectronic effects to control proline conformation and the electronic effects of aromatic rings to control the interactions between aromatic rings and proline. We are applying these approaches to stabilize protein secondary structures in peptides, to stabilize protein structures, to develop novel inhibitors of protein-protein interactions, and to design tunable proteins which are able to act conditionally, producing one structure or response in one environment and producing a contrary structure or response in a different environment.
Preprints
Pandey, A. K.; Ganguly, H. K.; Sinha, S. K.; Daniels, K. E.; Yap, G. P. A.; Patel, S.; Zondlo, N. J. "An Inherent Structural Difference Between Serine and Threonine Phosphorylation: Phosphothreonine Prefers an Ordered, Compact, Cyclic Conformation," BioRxiv 2020, DOI: 10.1101/2020.02.29.971382
Publications
Tressler, C. M.; Zondlo, N. J. "Perfluoro-tert-butyl Hydroxyprolines as Sensitive, Conformationally Responsive Molecular Probes: Detection of Protein Kinase Activity by 19F NMR," ACS Chemical Biology 2020, 15, 1096-1103. DOI: 10.1021/acschembio.0c00131
Urmey, A. R.; Zondlo, N. J. "Cysteine Oxidation to the Sulfinic Acid Induces Oxoform-Specific Lanthanide Binding and Fluorescence in a Designed Peptide," Free Radical Biology and Medicine 2020, 152, 166-174. DOI: 10.1016/j.freeradbiomed.2020.02.020
Urmey, A. R.; Zondlo, N. J. "Structural Preferences of Cysteine Sulfinic Acid: the Sulfinate Engages in Multiple Local Interactions with the Peptide Backbone," Free Radical Biology and Medicine 2020, 148, 96-107. DOI: 10.1016/j.freeradbiomed.2019.12.030
Zondlo, N. J. "SAR by 1D NMR," J. Med. Chem. 2019, 62, 9415-9417. DOI: 10.1021/acs.jmedchem.9b01688
Urmey, A. R.; Zondlo, N. J. "Synthesis of Peptides with Cysteine Sulfinic Acid via the Cysteine Methoxybenzyl Sulfone," Peptide Science 2019, e24137. DOI: 10.1002/pep2.24137
Costantini, N. V.; Ganguly, H. K.; Martin, M. I.; Wenzell, N. A.; Yap, G. P. A.; Zondlo, N. J. "T�h�e� �d�i�s�t�i�n�c�t� �c�o�n�f�o�r�m�a�t�i�o�n�a�l� �l�a�n�d�s�c�a�p�e�s� �o�f� �4�S�-s�u�b�s�t�i�t�u�t�e�d� �p�r�o�l�i�n�e�s� �t�h�a�t� �p�r�o�m�o�t�e� �a�n� �e�n�d�o� �r�i�n�g� �p�u�c�k�e�r�," Chem. Eur. J. 2019, 25, 11356-11364. DOI: 10.1002/chem.201902382
This paper was selected as a Hot Paper.
Urmey, A. R.; Zondlo, N. J. "Design of a Protein Motif Responsive to Tyrosine Nitration and an Encoded Turn-Off Sensor of Tyrosine Nitration," Biochemistry 2019, 58, 2822-2833. DOI: 10.1021/acs.biochem.9b00334
Gao, F.; Thornley, B. S.; Tressler, C. M.; Naduthambi, D.; Zondlo, N. J. "Phosphorylation-Dependent Protein Design: Design of a Minimal Protein Kinase-Inducible Domain," Org. Biomol. Chem. 2019, 17, 3984-3995. DOI: 10.1039/c9ob00502a
Wenzell, N. A.; Ganguly, H. K.; Pandey, A. K.; Bhatt, M. R.; Yap, G. P. A.; Zondlo, N. J. "E�l�e�c�t�r�o�n�i�c� �a�n�d� �s�t�e�r�i�c� �c�o�n�t�r�o�l� �o�f� �n�->π*� �i�n�t�e�r�a�c�t�i�o�n�s�:� �s�t�a�b�i�l�i�z�a�t�i�o�n� �o�f� �t�h�e� �α-h�e�l�i�x� �c�o�n�f�o�r�m�a�t�i�o�n� �w�i�t�h�o�u�t� �a� �h�y�d�r�o�g�e�n� �b�o�n�d�," ChemBioChem 2019, 20, 963-967. DOI: 10.1002/cbic.201800785
Scheuermann, M. J.; Forbes, C. R.; Zondlo, N. J. "Redox-Responsive Protein Design: Design of a Small Protein Motif Dependent on Glutathionylation," Biochemistry 2018, 57, 6956-6963. DOI: 10.1021/acs.biochem.8b00973
Tressler, C. M.; Zondlo, N. J. "Perfluoro-tert-butyl Homoserine is a Helix-Promoting, Highly Fluorinated, NMR-Sensitive Aliphatic Amino Acid: Detection of the Estrogen Receptor-Coactivator Protein-Protein Interaction by 19F NMR," Biochemistry 2017, 56, 1062-1074. DOI: 10.1021/acs.biochem.6b01020
Forbes, C. R.; Sinha, S. K.; Ganguly, H. K.; Bai, S.; Yap, G. P. A.; Patel, S.; Zondlo, N. J. "Insights into Thiol-Aromatic Interactions: A Stereoelectronic Basis for S-H/π Interactions," J. Am. Chem. Soc. 2017, 139, 1842-1855. DOI: 10.1021/jacs.6b08415
Tressler, C. M.; Zondlo, N. J. "Synthesis of Perfluoro-tert-butyl Tyrosine, for Applications in 19F NMR, via a Diazonium Coupling Reaction," Organic Letters 2016, 18, 6240-6243. DOI: 10.1021/acs.orglett.6b02858
Forbes, C. R.; Pandey, A. K.; Ganguly, H. K.; Yap, G. P. A.; Zondlo, N. J. "4R- and 4S-Iodophenyl Hydroxyproline, 4R-Pentynoyl Hydroxyproline, and S-Propargyl-4-Thiolphenylalanine: Conformationally Biased and Tunable Amino Acids for Bioorthogonal Reactions," Org. Biomol. Chem. 2016, 14, 2327-2346. DOI: 10.1039/c5ob02473k
Pandey, A. K.; Thomas, K. M.; Forbes, C. R.; Zondlo, N. J. "Tunable Control of Polyproline Helix (PPII) Structure via Aromatic Electronic Effects: An Electronic Switch of Polyproline Helix," Biochemistry 2014, 53, 5307-5314. DOI: 10.1021/bi500696k
Tressler, C. M.; Zondlo, N. J. "(2S,4R)- and (2S,4S)-Perfluoro-tert-butyl 4-Hydroxyproline: Two Conformationally Distinct Proline Amino Acids for Sensitive Application in 19F NMR," J. Org. Chem. 2014, 79, 5880-5886. DOI: 10.1021/jo5008674
Pandey, A. K.; Yap, G. P. A.; Zondlo, N. J. "(2S,4R)-4-Hydroxyproline(4-Nitrobenzoate): Strong Induction of Stereoelectronic Effects via a Readily Synthesized Proline Derivative. Crystallographic Observation of a Correlation between Torsion Angle and Bond Length in a Hyperconjugative Interaction," J. Org. Chem. 2014, 79, 4174-4179. DOI: 10.1021/jo500367d
Elbaum, M. B.; Zondlo, N. J. "OGlcNAcylation and Phosphorylation Have Similar Structural Effects in α-Helices: Post-Translational Modifications as Inducible Start and Stop Signals in α-Helices, with Greater Structural Effects on Threonine Modification," Biochemistry 2014, 53, 2242-2260. DOI: 10.1021/bi500117c
Brister, M. A.; Pandey, A. K.; Bielska, A. A.; Zondlo, N. J. "OGlcNAcylation and Phosphorylation Have Opposing Structural Effects in tau: Phosphothreonine Induces Particular Conformational Order," J. Am. Chem. Soc. 2014, 136, 3803-3816. DOI: 10.1021/ja407156m
This paper was chosen as a JACS Spotlight article: "Sorting Out How Structure Affects Function"
Naduthambi, D.; Bhor, S.; Elbaum, M. B.; Zondlo, N. J. "Synthesis of a Tetrasubstituted Tetrahydronaphthalene Scaffold for α-Helix Mimicry via a MgBr2-Catalyzed Friedel-Crafts Epoxide Cycloalkylation," Organic Letters 2013, 15, 4892-4895. DOI: 10.1021/ol402334j
Pandey, A. K.; Naduthambi, D.; Thomas, K. M.; Zondlo, N. J. "Proline Editing: A General and Practical Approach to the Synthesis of Functionally and Structurally Diverse Peptides. Analysis of Steric versus Stereoelectronic Effects of 4-Substituted Prolines on Conformation within Peptides," J. Am. Chem. Soc. 2013, 135, 4333-4363. DOI: 10.1021/ja3109664
Zondlo, N. J. "Aromatic-Proline Interactions: Electronically Tunable CH/π Interactions," Acc. Chem. Res. 2013, 46, 1039-1049. DOI: 10.1021/ar300087y
Brown, A. M.; Zondlo, N. J. "A Propensity Scale for Type II Polyproline Helices (PPII): Aromatic Amino Acids in Proline-Rich Sequences Strongly Disfavor PPII Due to Proline-Aromatic Interactions," Biochemistry 2012, 51, 5041-5051. DOI: 10.1021/bi3002924
Forbes, C. R.; Zondlo, N. J. "Synthesis of Thiophenylalanine-containing Peptides via Cu(I)-mediated Cross-Coupling," Organic Letters 2012, 14, 464-467. DOI: 10.1021/ol202947f
Balakrishnan, S.; Scheuermann, M. J.; Zondlo, N. J. "Arginine Mimetics via α-Guanidino Acids: Introduction of Functional Groups and Stereochemistry Adjacent to Recognition Guanidiniums in Peptides," ChemBioChem 2012, 13, 259-270. DOI: 10.1002/cbic.201100638
Zondlo, N. J. "Fold Globally, Bond Locally," Nature Chemical Biology 2010, 6, 567-568. DOI: 10.1038/nchembio.413
am Ende, C. W.; Meng, H. Y.; Ye, M.; Pandey, A. K.; Zondlo, N. J. "Design of Lanthanide Fingers: Compact Lanthanide-Binding Metalloproteins," ChemBioChem 2010, 11, 1738-1747. DOI: 10.1002/cbic.201000056
Zondlo, S. C.; Gao, F.; Zondlo, N. J. "Design of an Encodable Tyrosine Kinase-Inducible Domain: Detection of Tyrosine Kinase Activity by Terbium Luminescence," J. Am. Chem. Soc. 2010, 132, 5619-5621. DOI: 10.1021/ja100862u
Balakrishnan, S.; Zhao, C.; Zondlo, N. J. "Convergent and Stereospecific Synthesis of Molecules Containing α-Functionalized Guanidiniums via α-Guanidino Acids," J. Org. Chem. 2007, 72, 9834-9837. DOI: 10.1021/jo701766c
Naduthambi, D.; Zondlo, N. J. "Stereoelectronic Tuning of the Structure and Stability of the trp cage Miniprotein," J. Am. Chem. Soc. 2006, 128, 12430-12431. DOI: 10.1021/ja0648458
Zondlo, S. C.; Lee, A. E.; Zondlo, N. J. "Determinants of Specificity of MDM2 for the Activation Domains of p53 and p65: Proline27 Disrupts the MDM2-binding motif of p53," Biochemistry 2006, 45, 11945-11957. DOI: 10.1021/bi060309g
Balakrishnan, S.; Zondlo, N. J. "Design of a Protein Kinase-Inducible Domain," J. Am. Chem. Soc. 2006, 128, 5590-5591. DOI: 10.1021/ja057692h
Bielska, A. A.; Zondlo, N. J. "Hyperphosphorylation of tau Induces Local Polyproline II Helix," Biochemistry 2006, 45, 5527-5537. DOI: 10.1021/bi052662c
Thomas, K. M.; Naduthambi, D.; Zondlo, N. J. "Electronic Control of Amide cis-trans Isomerism via the Aromatic-Prolyl Interaction," J. Am. Chem. Soc. 2006, 128, 2216-2217. DOI: 10.1021/ja057901y
Meng, H. Y.; Thomas, K. M.; Lee, A. E.; Zondlo, N. J. "Effects of i and i+3 residue identity on cis-trans isomerism of the aromatici+1-prolyli+2 amide bond: Implications for Type VI β-turn Formation," Biopolymers (Peptide Science) 2006, 84, 192-204. DOI: 10.1002/bip.20382
Thomas, K. M.; Naduthambi, D.; Tririya, G.; Zondlo, N. J. "Proline Editing: A Divergent Strategy for the Synthesis of Conformationally Diverse Peptides," Organic Letters 2005, 7, 2397-2400. DOI: 10.1021/ol0506720

Group Members
Neal Zondlo, Principal Investigator.
Megh Bhatt, Graduate Student. B.S., M.S. Tribhuvan University
Himal Ganguly, Postdoctoral Fellow, Ph.D. Bose Institute
Greg Lada, Graduate Student. B.S., M.S. Juniata College
Aaron Lee, Graduate Student. B.S. Shepherd University
Brice Ludwig, Graduate Student. B.S. SUNY-Geneseo
Drew Urmey, Graduate Student. B.S. UMBC
Siyuan Xiang, Undergraduate Researcher, University of Delaware
Group Alumni
Shalini Balakrishnan, Ph.D. 2007 (Sloan Kettering)
Santosh Bhor, postdoctoral fellow 2006-2009 (Senior Scientist, ChemBioTek)
Agata Bielska, B.S. 2006 (M.D.-Ph.D., Washington University: Ph.D. in Biochemistry, 2013; Weill-Cornell; Sloan-Kettering)
Michael Brister, B.S. 2012 (GSK 2012-2013; M.D., Temple University, 2017; Thomas Jefferson University)
Alaina Brown, B.S. 2004 (M.D., University of Virginia, 2008; fellowship, Duke University; private practice)
Chris am Ende, B.S. 2005 (Ph.D., SUNY-Stony Brook, 2013) (Senior Scientist, Pfizer)
Kelly Daniels, B.S. 2016 (M.D. student, Thomas Jefferson University)
Kyle Davis, B.S. 2010 (Ph.D. student, Department of Environmental Sciences, University of Virginia)
Michael Elbaum, M.S. 2014 (Scientist, Medicinal Chemistry, Novartis Pharmaceuticals)
Christina Forbes, Ph.D. 2016 (post-doctoral fellow, Arizona State University)
Feng Gao, Ph.D. 2010 (Senior Scientist, Asymchem)
Rachel Jung, B.S. 2015 (Scientist, GlaxoSmithKline)
Jake Lefler, B.S. 2014 (Ph.D. student, Virginia Tech)
Kara Martin, B.S. 2013 (Ph.D. student, U Mass-Amherst)
Jennifer McCord, B.S. 2014 (Ph.D. student, Northeastern University)
Hai Yun Meng, M.S. 2006 (GenOn Energy)
Devan Naduthambi, postdoctoral fellow 2004-2008 (Senior Research Scientist, Gilead Sciences)
Anil Pandey, Ph.D. 2014 (Senior Scientist, Medicinal Chemistry, Aurigene Discovery Technologies)
Ryann Perez, B.S. 2020 (Ph.D. student, University of Pennsylvania)
Monica Pirigyi, B.S. 2013 (Associate Investigator, DuPont)
Nicole Raniszewski, B.S. 2019 (Ph.D. student, University of Pennsylvania
Rebecca Salomon, B.S. 2005 (Ph.D., University of Virginia, 2010) (Senior Scientist, Department of Defense)
Michael Scheuermann, M.S. 2011 (Cytec Engineered Materials)
Krista Thomas, M.S. 2005 (Associate Professor, Johnson County Community College)
Cay Tressler, Ph.D. 2017 (post-doctoral fellow, Johns Hopkins University)
Gasirat Tririya, postdoctoral fellow 2003-2005 (senior scientist, ASDI)
Genevieve Weist, B.S. 2016 (Ph.D. student, University of Pittsburgh)
Nicole Wenzell, B. S. 2017 (Ph.D. student, UCSF)
Mao Ye, postdoctoral fellow 2003-2004 (Assistant Director, Bioscience Synthetic Chemistry Core Facility, California NanoSystems Institute, UCLA)
Chen Zhao, M.S. 2005 (scientist, Wilmington Pharmatech)
Susan Carr Zondlo, senior research fellow 2003-2006 (Director of Bioanalysis, Department of Translational Medicine, QPS)
Lab Funding
NSF CHE (2020-2023)
NIH NIGMS (2019-2021)
NSF BIO MCB (2016-2020)
DOD CDMRP PRARP Alzheimer's Disease Research Program (2015-2020)
NIH NIAID (2015-2019)
NSF CBET (2014-2018)
NIH NIGMS (2010-2017)
National Science Foundation CAREER (2006-2011)
NIH COBRE
Alzheimer's Association
American Heart Association
National Science Foundation and the Intelligence Community
Undergraduate research funding: HHMI, Beckman Institute
Neal Zondlo Background
B. A. Rice University, summa cum laude, 1992. Biochemistry and Russian. (Undergraduate Research Advisors: Michael Stern (Rice), Richard L. Schowen (University of Kansas))
NSF Predoctoral fellow, Yale University, 1992-1995.
Ph. D. Yale University, 1999. (Graduate Advisor: Alanna Schepartz)
NIH postdoctoral fellow, Harvard University, 1999-2001. (Postdoctoral Advisor: Eric N. Jacobsen)
Contact Neal Zondlo
-
Mail:
-
Neal J. Zondlo
Professor
310 Drake Laboratory
Department of Chemistry and Biochemistry
University of Delaware
Newark, DE 19716
Phone: 302-831-0197 (please do not leave voicemail: instead contact by email)
Fax: 302-831-6335
Email: zondlo@udel.edu (preferred contact)

 The complexity of humans is dependent on post-translational modifications of proteins. The most common post-translational modification is the phosphorylation of serine, threonine, and tyrosine residues by protein kinases. Protein kinases are tightly regulated, and changes in kinase activity are associated with most human diseases, including cancer, heart disease, and Alzheimer's disease. We are developing approaches to understand how protein phosphorylation changes the structure of proteins toward understanding the mechanisms associating changes in kinase activity with human diseases. In addition, we are developing new tools to understand the changes in kinase activity associated with human disease. We have designed new protein structures, called protein kinase-inducible domains, whose structures are dependent on their phosphorylation state. These designed proteins are under the control of specific protein kinases and are non-fluorescent when not phosphorylated, but highly fluorescent when phosphorylated, and may act as genetically encoded sensors of protein kinase activity.
The complexity of humans is dependent on post-translational modifications of proteins. The most common post-translational modification is the phosphorylation of serine, threonine, and tyrosine residues by protein kinases. Protein kinases are tightly regulated, and changes in kinase activity are associated with most human diseases, including cancer, heart disease, and Alzheimer's disease. We are developing approaches to understand how protein phosphorylation changes the structure of proteins toward understanding the mechanisms associating changes in kinase activity with human diseases. In addition, we are developing new tools to understand the changes in kinase activity associated with human disease. We have designed new protein structures, called protein kinase-inducible domains, whose structures are dependent on their phosphorylation state. These designed proteins are under the control of specific protein kinases and are non-fluorescent when not phosphorylated, but highly fluorescent when phosphorylated, and may act as genetically encoded sensors of protein kinase activity.
 Work in genomics and proteomics is revealing vast numbers of interaction loci within the cellular milieu, and thus vast numbers of potential targets for agonists and antagonists of protein-protein, protein-DNA and protein-RNA interactions. Our focus is the use of molecular design and organic synthesis to develop small molecules which mimic larger biological structures. Our work involves the development of appropriate, readily accessible organic scaffolds, in solution and on solid phase, using modern methods of organic synthesis. To evaluate our scaffolds they are subjected to high-throughput testing for biological activity. Modularity in synthesis allows the combination of multiple structural elements to allow recognition of larger protein surfaces and the synthesis of multifunctional "proteins." The post-genomic era requires novel tools to elucidate the identity, classes and modes of protein-protein interactions in disparate cell types, developmental stages, and intracellular environments, in addition to changes due to age and disease states. The ability to generate small molecule mimics of protein recognition elements permits their use as chemical probes of protein-protein interactions.
Work in genomics and proteomics is revealing vast numbers of interaction loci within the cellular milieu, and thus vast numbers of potential targets for agonists and antagonists of protein-protein, protein-DNA and protein-RNA interactions. Our focus is the use of molecular design and organic synthesis to develop small molecules which mimic larger biological structures. Our work involves the development of appropriate, readily accessible organic scaffolds, in solution and on solid phase, using modern methods of organic synthesis. To evaluate our scaffolds they are subjected to high-throughput testing for biological activity. Modularity in synthesis allows the combination of multiple structural elements to allow recognition of larger protein surfaces and the synthesis of multifunctional "proteins." The post-genomic era requires novel tools to elucidate the identity, classes and modes of protein-protein interactions in disparate cell types, developmental stages, and intracellular environments, in addition to changes due to age and disease states. The ability to generate small molecule mimics of protein recognition elements permits their use as chemical probes of protein-protein interactions.





